Antibiotic Resistance: blaNDM, mcr-1 and mcr-2, The Blacklisted Genes
Carole Mansour#, Katia Mansour#, Jameel Kobeissi, Bassam Hamam, Roland Bou Raad, Lea Saliba, Imtithal Sheet and Samer Sakr*
Department of Biological and Chemical Sciences, School of Arts and Sciences, Lebanese International University, Lebanon
# - Authors with equal contribution to this work
Received Date: 12/06/2020; Published Date: 02/07/2020
*Corresponding author: Samer Sakr, School of Arts and Sciences, Lebanese International University, Beirut, Lebanon. P.O. Box 146404; Tel: 961-1-706881; Email: samer.sakr@liu.edu.lb
Abstract
The emergence and spread of Antibiotic Resistance Genes (ARG) into multidrug resistant bacteria have become a global health threat. Resistance to carbapenem, the last line of antibiotic, encoded by the plasmid-borne gene blaNDM, have been reported in China in 2009 and was described as a problem of growing significance due to limited treatment options. Difficulty in treating infections caused by carbapenem resistant species led to the renewal use of polymyxin compounds such as colistin. Reconsidering colistin as a last line resort antibiotic for treatment of carbapenem-resistance-Gram-negative infections resulted in the extensive use of the drug. Unfortunately, mobilized colistin resistance encoded by plasmid-borne gene mcr-1 was identified in China in 2015. This was followed by the discovery of mcr-2, colistin resistant gene distinct from mcr-1, with the ability to confer high spread frequency among closely and distantly related species. This review explores the prevalence, spread, aggregation and mechanisms of resistance of two genes that are challenging all available lifesaving therapeutics; blaNDM and mcr-1 and mcr-2.
Keywords: Antibiotic resistance; Gram-negative; Carbapenem; blaNDM; Colistin; mcr-1; mcr-2
Introduction
Bacteria, one of the most diverse kingdoms of microorganisms, are amongst the oldest and structurally simplest forms of life on earth. These unicellular living organisms are characterized by the presence of a cell wall around their cell membrane [1]. Bacterial cell wall is essential for the maintenance of the shape and the integrity of the cell, thus protecting it from damage such as swelling or rupturing [2]. The cell wall is made up of peptidoglycan (PPG), which consists of a polysaccharide network connected via polypeptides cross linkages [3]. In some bacteria, the peptidoglycan is found around the outer surface of the cell forming a thick and complex network, in other bacteria the peptidoglycan forms a thin layer between two plasma membranes [4]. The two major types of bacteria were distinguished using the Gram staining technique; Gram-positive bacteria were reported to have a thicker peptidoglycan compared to compared to a thin peptidoglycan layer in gram negative bacteria [5]. The thin peptidoglycan layer in Gram-negative bacteria is located between the plasma membrane and an outer membrane. The latter is composed of phospholipids concentrated on the inner leaflet whereas the outer leaflet contains mainly glycolipids and lipopolysaccharides (LPS or endotoxin). Moreover, the LPS is a unique component of the outer surface and is made up of a specific carbohydrate-lipid moiety termed lipid A [6]. Lipid A is attached to a linear polysaccharide region consisting of a core and an O-specific polysaccharide [6, 7]. In fact, LPS is responsible of eliciting strong immune responses in various animal species with the lipid A being the primary immunity stimulator center [7]. In order to treat infections caused by pathogenic bacteria, the most commonly used family of drugs is called antibiotics. These antimicrobial agents are capable of killing bacteria, or stopping their growth. Depending on the range of their targets, antibiotics are classified into two types: narrow and broad-spectrum. The term broad-spectrum antibiotic refers to an antibiotic that acts against a wide range of disease-causing bacteria [8]. It acts against both Gram-positive and Gram-negative bacteria in contrast to a narrow-spectrum antibiotic, which is only active against specific families [9]. Since their introduction in the early 1940s, antibiotics have been a turning point in the history of medicine. However, studies from across the world have shown that some bacteria have developed resistance to commonly used antibiotics, leading to the loss of the potency and the efficiency of this class of drugs. Despite all the extensive research and major breakthroughs in antibiotic development and treatment advancements, recent evidence revealed the emergence of resistance to all first-line and last-resort antibiotics [10]. The molecular mechanism of antibiotic resistance development exhibited by Different Multidrug Resistant (MDR) bacteria has been central to extensive studies. A variety of genomic alterations can be the root of antibiotic resistance acquisition and studies showed that Mobile Genetic Elements (MGEs) are key factors in enabling resistance transfer mechanism among bacterial population to survive stress conditions [11]. Recently, extreme concerns have increased with the emergence of MGEs carrying genes encoding enzymes for Extended Spectrum β-Lactamases (ESBLs) carbapenems. Rapid spread of carbapenemases was reported earlier after New Delhi Metallo β-lactamase-1 (NDM-1) was identified in 2008 in Sweden from a patient who was previously hospitalized in India [12]. Emergence of blaNDM resistant gene encoding NDM-1 is now the focus of a global attention since it is reported from all continents; most isolates have shown failure to treatment to all available antibiotics [13]. Another more recent burden is the emergence of the first transferable colistin antibiotic resistance gene, mcr-1 which was identified in Enterobacteriaceae – a species of bacteria that mainly inhabits animals’ gastrointestinal tract (GIT) - mainly in Escherichia coli. Since then, mcr-1 gene has been detected/reported worldwide [14]. In 2016, a new gene termed mcr-2 gene was identified in Belgium and its encoded polypeptide was described to share 80.6% amino acid identity with MCR-1 [15]. The aim of this report is to present an overview of the current knowledge on blaNDM and mcr resistant genes and their variants, and to review recent developments in our understanding of their prevalence worldwide.
Understanding Resistance
The emergence of antibiotic resistant bacteria has shown a significant rise in the past decade. It has been widely documented that the use of antibiotics leads to dissemination and amplification of resistant genes via natural selection [16]. Natural selection processes in diseases-causing bacteria have been studied extensively. Frequently used antimicrobial drugs have accelerated the pace of advent of selection among bacterial species; this increased selective pressure in bacterial population allowed strains to evolve robust mechanisms to bypass the effective action of antibiotics and become resistant [17]. Resistance appearance due to selective pressure represents a Darwinian competition among bacterial species [18]. In fact, recent studies involving meta-genomic analysis of soil pathogens revealed widespread variance of Antibiotic Resistant Genes (ARGs) of which only a fragment have been identified in human pathogens [19]. In addition to selection of antimicrobial resistance through mutations in genes encoded on a microbe’s chromosome, new genetic material can also be exchanged between organisms. Thus, bacteria develop resistance by (1) Random mutations and Vertical Gene Transfer (VGT) - the transmission of DNA from parent to offspring and (2) Exchange of genetic material via Horizontal Gene Transfer (HGT) – the transmission of DNA between microorganisms and the expression of these transferred ARGs [20]. Currently, most studies describe mechanisms of resistance acquisition and dissemination as a result of HGT rather than mutational events, with the former to be considered as the most important factor in the current outbreak of Anti-Microbial Resistance (AMR) [21]. Horizontal gene transfer is a process that involves the transfer of Mobile Genetics Elements (MBE) between strains that are either closely or distantly related. The three major mechanisms by which bacteria transfer genes horizontally are Conjugation, Natural Transformation, and Transduction [22]; it has been previously shown that the main mechanism of HGT is through conjugation [23]. Gene transfer via conjugation has been studied extensively and the frequencies of the transfer events showed significant variance. Experimental data suggest that frequencies of conjugative events in nature are presumably several orders of magnitude higher than those under laboratory conditions [24]. Conjugation is the transfer of DNA through a multi-step process via cell surface pili or adhesins. It is promoted by the conjugative machinery which is encoded either by genes on autonomously replicating plasmids or by Integrative Conjugative Elements (ICEs) in the chromosome [25]. However, many of the genes that enable bacteria to metabolize organic compounds that are toxic to them, such as antibiotics, are carried by plasmids [26,27]. Typically, plasmids are extra chromosomal DNA carrying genes required for replication machinery. In addition to their function in replication, studies have showed that plasmids are heavily implicated in the transfer of resistance genes [28, 29]. Plasmid genome consists of “core” and “accessory” genes. Core genes are involved mainly in plasmid transmission and replication, whereas accessory genes function in increasing bacterial fitness and survival under certain environmental stress conditions [30, 31]. In fact, antibiotic resistance in many pathogenic bacteria has evolved by acquiring plasmids carrying antibiotic resistant genes – such as blaNDM and mcr-1 and mcr-2.– that are derived from either closely or distantly related bacteria [32, 33]. In this context, some plasmids have a broad host range and can transfer between different species whereas others exhibit much narrower effect and thus are confined to one genus or species [34].
Beta Lactams
Discovery of β-Lactams; A Brief History
Penicillin: It all began in the 1920s when Sir Alexander Fleming, a Scottish bacteriologist, was studying the relationship between colony morphology and virulence in Staphylococcal species. Fleming noticed that some of the cultures were contaminated with molds. Colonies in proximity to mold were undergoing lysis. The dissolving and transparent surrounding agar gel supported his observation [35]. Upon subsequent experiments on several bacterial cultures, Fleming was able to confirm that the fungal extract exhibited an “inhibitory” property against most Gram-positive bacteria such as Staphylococcus aureus, Bacillus anthracis, Streptococcus pyogenes, Corynebacterium diphtheria, but not against Gram-negative bacteria such as Escherichia coli, Klebsiella pneumonia, Vibrio cholerae and others. The fungal extract was termed “Penicillin”, since it was derived from Penicillium notatum mold [36]. However, Fleming found it difficult to extract and isolate his so-called “mold juice” in large quantities. In fact, it was only until 1940, that Penicillin was transformed from a laboratory interest into a life-saving drug during World War II when Howard Florey, Ernst Chain and their colleagues at the Oxford University were able to isolate and mass produce the drug [37]. The initial recognition of Penicillin and its clinical introduction was recognized as one of the greatest advances in the history of medicine.
β-Lactam Structure: The Building Block: Further interest in the activity of the compound revealed that different strains, culture conditions, and media result in the production of different penicillin forms [38]. Two forms of the extract were used at that time: 2-pentenyl penicillin and benzyl-penicillin, commonly known as Penicillin F or I and Penicillin G or II, respectively. In fact, Florey and his colleagues proceeded to crystallize and identify the structure of the compound; the two forms were found to share a common feature with differences only in their R group [39]. The common feature was a four-membered lactam, a cyclic amide, and was termed β-lactam because the nitrogen atom is attached to the β-carbon atom relative to the carbonyl (C=O) group [40]. The ring is further fused into 5- or 6- membered rings depending on the group of β-lactam.
β-Lactam Derivatives: The discovery of Penicillin and its successful introduction into clinical practice led to the robust search for other antibiotics and natural sources continued to be explored. Novel β-lactam compounds were developed and modified continuously and hundreds of β-lactam antibiotics have been developed (naturally occurring or semi-synthetic). Members of β-lactam antibiotics of clinical importance include the penicillin, cephalosporins, monobactams and carbapenems [41, 42]. The penicillins and cephalosporins contain the β-lactam ring fused to 5- and 6-membered rings, respectively, which contain a carboxyl group at the C-3 and C-4 positions. As a group, these antibiotics display a wide range of activity against Gram-positive and Gram-negative bacteria [41]. In contrast, monobactams do not contain a fused ring structure. They consist of the β -lactam ring linked to a sulfonic acid group at the analogous position of the carboxylate group found in previously described penicillins and cephalosporins. Moreover, monobactam antibiotics are active against aerobic Gram-negative bacteria [43].
Finally, carbapenem β-lactam members possess the broadest spectrum of activity and greatest potency against Gram-positive and Gram-negative bacteria. As a result, carbapenems are often used as “last-line” or “antibiotics of last resort” when patients with bacterial infections fail to respond to classical treatment or when infection is suspected to involve resistant bacteria [44]. In the next section of this review, we describe the current state of carbapenem family of β-lactams with an emphasis on the landscape of carbapenem resistance development.
Mechanism of Action of β-Lactams
Overview of Bacterial Cell Wall Biosynthesis: To grow and divide efficiently, bacterial cells must add new peptidoglycan to its wall. The peptidoglycan layer consists of a network of alternating polysaccharides, N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG), with pentapeptide chains cross-linking NAM residues of neighboring chains. Peptidoglycan synthesis is divided into three stages that occur at three different locations in the cell [45]. The first stage takes place in the cytoplasm and involves the synthesis of the nucleotide sugar-linked precursors UDP-N-acetylmuramyl (UDP-MurNAc)-pentapeptide and UDP-N-acetylglucosamine (UDP-GlcNAc) [46]. During the second stage, which takes place in the cytoplasmic membrane, precursor lipid intermediates are produced (specifically, lipid I and lipid II) [46, 47]. The final stage occurs at the outer part of the cytoplasmic membrane. It involves the polymerization of the newly synthesized disaccharide-peptide unit and its addition to the nascent peptidoglycan [48]. This is carried out mainly by the action of a specific class of enzymes called penicillin binding proteins, or PBP [49, 50]. PBP enzymes are responsible of cross-linking the peptidoglycan chains and include (i) transglycosylases that catalyze the formation of glycosidic bonds; (ii) transpeptidases that catalyze the formation of peptide bonds; and (iii) carboxypeptidases that hydrolyze peptide bonds at C-terminus [51]. PBPs were initially identified because of their ability to covalently bind penicillin. In addition, depending on the organism, the peptide chains are further modified [52] (Figure 1).
β-Lactams Activity Against Bacterial Wall: β-Lactam antibiotics block the biosynthesis of peptidoglycan wall by inhibiting PBPs, making them inefficient in achieving their role in cell wall synthesis. In fact, the structural similarity between β-Lactam antibiotics and D-alanyl-D-alanine – the terminal amino acid residue on NAM - facilitates their binding irreversibly to the active site of PBPs (a serine residue) [52, 53]. Subsequently, cross-linkage between adjacent NAMs will be prevented. This hindrance greatly affects and disrupts the structural integrity of bacterial cell wall, leading to osmotic instability and, eventually, driving the cell to undergo autolysis [54] (Figure 1).
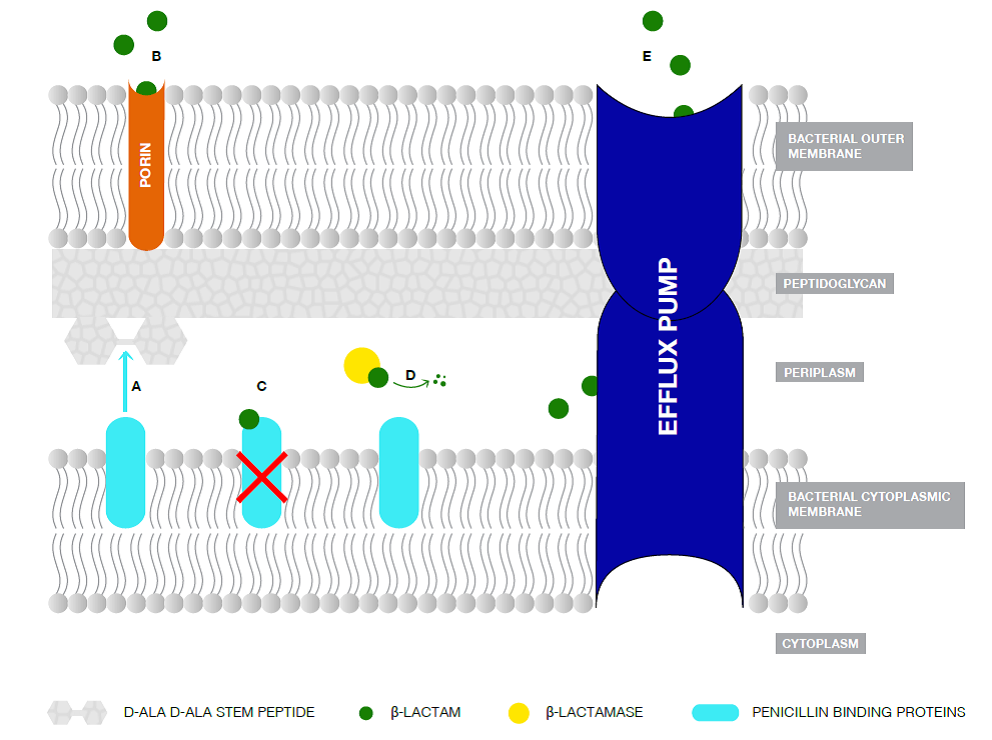
Figure 1: Schematic representation of the Gram-negative bacterial membrane structures, including the interaction of β-lactam antibiotics and β-lactamases [69].
A. Penicillin-Binding Proteins (PBPs) with transpeptidase activity are involved in cell wall biosynthesis by catalyzing the crosslinking of adjacent peptides at the terminal D-Ala-D-Ala portion of the peptidoglycan stem peptide. B. β-Lactams are transported to the periplasmic space via porins. C. β-lactams binds covalently to the transpeptidases forming a stable ester adduct, thus inhibiting them. D. When exported to the periplasm, β-Lactamase enzymes are attached to the inner leaflet of the bacterial outer membrane via an N-terminal lipid. They hydrolyze the β-lactam antibiotics preventing them from inhibiting the transpeptidases. E. Efflux pumps transports actively the antibiotics to the external environment avoiding their deadly effect on the bacterial cell
Mechanisms of Resistance to β-Lactams
Emergence of β-Lactamases: It is important to mention that the microbial genetic capacity to evolve and spread resistance was underestimated. In fact, resistance to penicillin was reported shortly after its introduction in 1945 [55]. This was followed by a critical number of cases reporting emergence of resistant species to β-lactam antibiotics [56]. Several mechanisms of bacterial resistance acquisition have been described and these mainly include: (i) decreased access of antimicrobials to the target PBPs (efflux pumps) [57]; (ii) alteration of the sequence of target transpeptidases by mutation or recombination to create enzymes that bind poorly to β-lactams; (iii) lowered permeability (decreasing the number of outer membrane porins thus restricting the influx of antibiotics); and (iv) hydrolysis and inactivation by enzymes termed β-lactamases [58] (Figure 2). Despite the significance of the previously described processes, the production of β-lactamase enzymes is the most common mechanism of bacterial resistance to β-lactam antibiotics [59].
The continuous exposure of bacterial strains to a multitude of β-lactams has led to the overproduction and mutation of β-lactamases among different species of bacteria.
Development of β-Lactamases: β-Lactamases are bacterial enzymes that hydrolytically inactivate the β-lactam family of antibiotics [60]. Following their appearance, extensive research focused on the development of extended-spectrum β-Lactam antibiotics. However, later in the 1980s, extended-spectrum β-lactamases (ESBLs) rapidly evolved in response to newer β-lactam antibiotics and they were termed so to reflect theexpanded substrate spectrum of these enzymes [61,62]. These enzymes were frequently reported in Klebsiella pneumoniae (K. pneumoniae), Escherichia coli (E. coli), Acinetobacter baumannii (A. baumannii), Pseudomonas aeruginosa (P. aeruginosa), Salmonella typhimurium (S. Typhimurium), Enterobacter species, Proteus species, Serratia species, among others. Moreover, genes encoding β-lactamases can be found on the bacterial chromosome or on plasmids; many studies described the HGT of these genes as the most common mechanism of acquiring resistance to this family of antibiotics [63].
Classification of β-Lactamases: Two current classification schemes for β-lactamases are globally accepted. The first one is based on the amino-acid sequence and the second one is based on the functionality; in this review we use the first classification system for the interest of its subclasses [64]. Based on the primary homology sequence, β-lactamases were divided into four classes: A, B, C, and D enzymes [65]. Classes A, C, and D are active-site serine enzymes that catalyze, via a serine-bound acyl intermediate, the hydrolysis of the β-lactam ring. They are termed serine-β-lactamases [66,67]. Class B enzymes, called metallo-β-lactamases (or simply metalloenzymes), require divalent zinc ions for their activity (substrate hydrolysis) and are further discussed later in this context [68, 69].
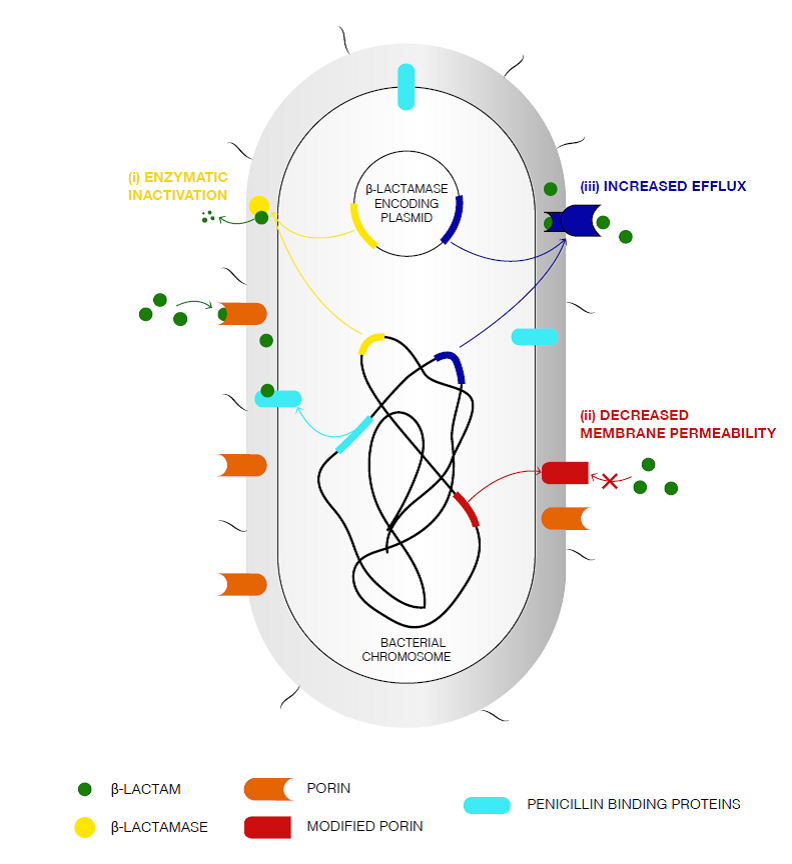
Figure 2: Primary mechanisms of ß-lactam resistance [25].
The porins are responsible of the uptake of the β-lactams through the bacterial outer membrane. PBPs binds irreversibly to the β-lactams in the periplasmic-interspace, leading to the inhibition of the peptidoglycan synthesis. Primary mechanisms of β -lactam resistance include the following: (i) enzymatic inactivation of the antibiotic by chromosome- and/or plasmid-encoded enzymes possessing hydrolytic activity against β -lactam molecules; (ii) decreased outer membrane permeability via: production of modified porins, downregulation of porin expression, or a shift in the types of porins found in the outer membrane; and (iii) Upregulation of the efflux pumps leading to the export of the antibiotic to the external medium.
Carbapenem
Overview
Following the emergence and widespread of β-lactamases and the threat over the usage of penicillin and its derivatives, the search for inhibitors began [70]. The first carbapenem was developed and introduced for clinical use in 1985. Carbapenems are part of the β-lactam class of antibiotics. They consist of a β-lactam ring fused to a 5-membered ring that has a carbon replacing the sulfur at C-1 and also containing a double bond between C-2 and C-3.
Mechanism of Action of Carbapenem
As a class of β-Lactam antibiotics, carbapenem acts by targeting cell wall biosynthesis via PBP inhibition. Interaction with different types of PBPs occurs on cell surface of Gram-positive bacteria and in periplasmic space of Gram-negative bacteria [71]. In the latter, carbapenems cannot diffuse easily through the bacterial cell wall, and therefore must enter through water filled outer membrane proteins known as porins. Following their entry, carbapenems behave similarly to other β-Lactams and “permanently” acylate PBPs rendering them inactive and eventually leading to bacterial cell death [72].
Resistance to Carbapenem
Emergence of Carbapenemases: Since described as the “last resort antibiotic”, the emergence and spread of acquired carbapenem resistance represents a major public health concern [73]. Resistance to carbapenems was first observed in enterobacterial isolates, especially among Enterobacter spp [74]. Recently, global spread of carbapenemase has increased significantly and is considered as a major global threat. Carbapenemases are defined as β-lactamases with activity against most β-lactams and significant activity against carbapenems [75].
Classification of Carbapenemases: Like the other beta-lactamases, carbapenemases are generally classified into functional groups and molecular classes. Following the molecular scheme produced by Ambler and colleagues, carbapenemases are divided into serine-carbapenemases (Class A, and D) and metallocarbapenamases (Class B, commonly called metallo-beta-lactamses or MBLs) [76, 77]. We focus in this review on Carbapenemases with the greatest clinical relevance worldwide, MBLs.
Metallocarbapenemases: The first MBLs were isolated from environmental and opportunistic bacteria such as Bacillus cereus [78, 79], Aeromonas spp. [80, 81], and Stenotrophomonas maltophilia [82]. Further Studies revealed that MBL genes of these soil dwelling pathogens were intrinsic and chromosome-borne. Since the 1990s, an increase in acquired MBL genes has been reported in Pseudomonas spp. [83], and Enterobacteriaceae [84; 85]. The most common families of the identified plasmid-mediated MBL genes mainly include IMP- [86], and VIM-type enzymes [87, 88], together with the emerging NDM-type [89, 90]. Reports showed that VIM and IMP-type enzymes have the largest numbers of enzyme derivatives. However, the greatest clinical impact has been correlated mainly to NDM-enzymes.
blaNDM: The Carbapenem Resistant Gene
Identification: In 2007, a 59-year-old Swedish male patient of Indian origin traveled to India and was diagnosed and hospitalized for gluteal abscess (a collection of pus under the skin close to the anus) [89]. A year after, he was sent back to a hospital in Sweden where a Urinary Tract Infection (UTI) was confirmed by obtaining a urine culture that was positive for Klebsiella pneumoniae showing resistance to antibiotics including carbapenems [89]. In March 2008, following transfer of the patient to a nursing home, test samples of stool showed a carbapenem resistant strain of E.coli [89]. Results of phenotypic tests of both isolates proposed that resistance was caused by the production of MBLs. Interestingly, none of the known genes coding for MBLs were detected by PCR analysis. In fact, after series of cloning and sequencing experiments, a novel type of enzyme was identified and was found to share 32% identity with the other MBLs [89]. This MBL was named NDM-1 and its gene blaNDM-1 after New Delhi, the capital city of India, by Yong et al. who believed that the resistance originated from there [89]. In 2010, in the United Kingdom, another case was reported with an infection of E.coli expressing NDM-1 [91]. The infected patient was of an Indian descent and had undergone dialysis 18 months previously while visiting India. Studies showed that the bacterium was fully resistant to all antibiotics except to the polymyxin antibiotic colistin. The fact that the same novel resistance gene was present in two different genera implied that it was transferable. Many strains thereafter were analyzed and revealed to be carrying blaNDM-1 on plasmids, allowing it to transfer between multiple strains of bacteria via horizontal gene transfer. Moreover, all of the samples were resistant to different classes of antibiotics but susceptible to colistin. Later in 2010, an environmental point prevalence study was conducted on samples collected from seepage samples and drinking water in New Delhi. Twenty strains of bacteria were found carrying the blaNDM-1 gene [92]. In August 10, the first death due to an infection with bacteria expressing the NDM-1 enzyme was reported in Belgium. The deceased was hospitalized with a major leg injury in Pakistan and was treated in a hospital there. He was then repatriated to Belgium and passed away despite the administration of colistin.
Infection Site: NDM-1 producing bacteria have been recovered from many infection sites. They have been found in patients with urinary tract infections, pneumonia, septicaemia, wound infections and device-associated infections [93, 94].
Global Dissemination: There has been an increase in the number of articles about the ‘New Delhi Metallo-betalactamase-1’ enzyme added to the PubMed database since 2010, but the current spread of NDM- 1-prodcucing bacteria in 2016 is likely broader than the published reports suggest. The rapid global spread of NDM-type carbapenemases may be partly attributed to the dissemination of various epidemic broad-host-range plasmids bearing the blaNDM genes. Since 2006, NDM- carrying bacteria have been reported from 40 countries covering all continents except South America and Antarctica. At present, most isolated bacteria have originated from people that were either infected or colonized on the Indian subcontinent and who have then traveled elsewhere [95, 96]. There are other reservoirs of colonized and/or infected patients in the Balkan countries and in the Middle East, where people often travel to and from the Indian subcontinent [97, 98].
Epidemiological link of NDM with the Indian subcontinent: An epidemiological link to the Indian subcontinent, the Balkans and the Middle East has been demonstrated in most of the reports of the global spread of NDM producers, identifying these geographic regions as endemic for NDM [99,100]. In particular, NDM-positive Enterobacteriaceae were found to be geographically widespread in the Indian subcontinent, being recovered from ten areas in India, eight areas in Pakistan and one area of Bangladesh. Many factors have influenced the geographically widespread emergence of NDM-1-producing bacteria: (i) the increase in long-distance travels [101], (ii) the increase in international travels to access medical care [102], and (iii) the widespread access to broad-spectrum antibiotics. Given the volume of global travel, the quality of hygienic standards in many countries, and the number of humans carrying NDM-1-producing bacteria, it is likely that these bacteria will continue to spread worldwide [94].
Bacterial Species Dissemination: The blaNDM-1 gene was reported in multiple Enterobacteriaceae sp., mainly in epidemic clones of K. pneumoniae and E. coli. Other Gram-negative bacteria, such as some strains of Vibrio cholerae, Pseudomonas spp. and A. baumannii were also reported producing NDM-1 [103, 104]. Retrospective Studies of cultures of Enterobacteriaceae isolates dating back to 2006 revealed the presence of blaNDM-1. Analyses of the genetic environment revealed that dissemination of blaNDM-1 might have originated from A. baumanni.
NDM- producing Klebsiella pneumonia: Klebsiella pneumoniae is the most important member of Klebsiella genus of the Enterobateriaceae family. In recent years, it became a key pathogen in global nosocomial dominance. It inhabits humans as well as the ecological environment, such as water, soil, and sewage. In addition, K. pneumoniae is capable of surviving in extreme environments for long periods of time [105]. K. pneumoniae producing NDMs have spread rapidly [105]. They are considered to be endemic in the Indian subcontinent, including India, Pakistan, and Bangladesh as mentioned previously [106]. In fact, NDM-1 was reported to be the most common carbapenemase type detected in India. It accounted for 75.22% of the carbapenemase-producing isolates [107]. The movement of patients between countries may be the reason for the international spread of carbapenemase-producing K. pneumoniae [108]. K. pneumoniae and other Gram-negative bacteria that produce carbapenemase were obtained from patients that had recently traveled outside Canada between the years 2010 and 2013. These isolates were found to be NDM-1 producing K. pneumoniae that were imported from India to Canada [109]. Thus, appropriate attention and care are required in the treatment of patients with a history of hospitalization in foreign countries in which carbapenemase-producing bacteria are heavily reported and documented.
Structural features: As mentioned above, class B1 β-lactamases consists of 3 types, IMP, VIM, and the newly discovered NDM. Through reviewing articles reporting the occurrence of IMP, VIM or NDM, a global distribution map was developed by Maria F. Mojica and her colleagues. The authors noted that as of July 2014, the only country that reported only IMP and NDM was Lebanon. The representative B1 MBLs appear to have structurally conserved active-site features in the primary zinc ligands. Crystal structures of B1 β-lactamases showed a mononuclear or dinuclear metal ion cluster in the active site [110, 111]. Zn1 site is occupied by the sole zinc ion in monozinc B1 MBLs. Dizinc B1 MBLs structures fall in two categories. In the first, dizinc site binds both zinc ions with positive cooperativity, with binding of the first favoring the binding of the second [112]. In other structural studies of dizinc B1 MBLs with differing affinities for each site, Zn1 is refined with higher occupancy than Zn2 implying that Zn2 to be the weaker binding site [113, 114]. Some crystal structures of the B1 β-lactamase NDM-1 a third zinc ion (Zn3) bound at the edge of the active site [115].
Colistin
Overview
Following their discovery in the 1940s, the polymyxin group of polypeptide antibiotics showed effective therapeutic activity in the treatment of infections caused by Gram-negative bacteria [116,117]. Groups of polymyxins are produced by Bacillus species and they consist of 5 chemically different compounds (polymyxins A–E), with only polymyxin B and polymyxin E, also known as PMB and Colistin respectively, commonly used in clinical practices [118]. PMB is produced by Bacillus polymyxa; while Colistin is produced by Bacillus colistinus [119].
However, administration of polymyxins was associated with reports on nephrotoxicity and neurotoxicity in the 1970s, and thus compounds of this class were gradually withdrawn and replaced with other antibiotics of less toxicity [120,121]. In the past two decades, the emergence of MDR pathogens, and the lack of development of new antibiotics with activity against them have threatened therapeutic choices. Subsequently, this led to the revival and reconsideration of the usage of this class of antibiotics in clinical practice; in fact colistin and PMB were considered to be the last resort for the treatment of infections caused by MDR Gram-negative bacteria [122, 123]. The current review focuses on colistin, rather than PMB, because of its widely described use clinically.
Chemical Structure
As mentioned earlier, Colistin is a polypeptide antibiotic of the polymyxin family. It is non-ribosomally synthesized with a molecular weight of 1750 Da. Polymyxins consist of a decapeptide (an oligopeptide containing ten amino acids) that comprises a cyclic heptapeptidecore (seven amino acids) linked to a linear tripeptide segment that holds an N-terminal fatty acyl tail [124,125]. The amino acid composition of the molecule is as follows: d-leucine, l-threonine, and l-a-g-diaminobutyric acid with the latter attached to a fatty acid group. Depending on the composition of the fatty acid residue, one can distinguish two classes: Colistin A with the fatty acid residue attached identified as 6-methyl-octan-oic acid, or 6-methyl-eptanoic acid for Colistin B [126; 127]. Moreover, two types of colistin are synthesized for clinical application; colistin sulfate in the form of sulfate salt; while the second is Colisti-Methate Sodium (CMS) in the form of sodium salt [128]. CMS – colistin derivative – is unstable both in vivo and in vitro. However, once in the plasma, the molecule undergoes a spontaneous hydrolysis reaction and releases the active form colistin [129,130]. The hydrophilic peptide chain, and the hydrophobic fatty acid portion give the molecule of colistin an overall amphipathic characteristic; this provides a stable reaction with polar and non-polar targets such as the previously described LPS membrane of the bacteria [131]. Next, we describe the specific mechanism by which colistin targets lipid A component of LPS.
Mechanism of Action
Colistin and formerly described polymyxin B are commonly active against MDR Gram-negative bacteria including P. aeruginosa, A. baumannii and K. pneumoniae that previously showed resistance to all current available antibiotic therapeutics [132,133]. The primary target of colistin is the polyanionic LPS component of the outer membrane. As described earlier, LPS of Gram-negative cell wall is made up of a double lipid layer and polysaccharides that contain negative charges that are believed to play major role in maintaining the integrity of the cell wall. Moreover, LPS components are further stabilized via bivalent cations such as calcium (Ca++) and magnesium (Mg++) found linked to phosphate groups of the LPS [134]. As mentioned previously, colistin molecules have a strong positive charge and a hydrophobic acyl chain that give them a high binding affinity to LPS molecules. In fact, having a positive charge is essential for polymyxins activity [135]. The interaction between colistin and the LPS is best described in a detergent-like fashion in a two-step process. Initially, binding of colistin to its target is facilitated through electrostatic interactions between the cat4ionic polypeptide and anionic LPS molecules in the outer membrane of the Gram-negative bacteria [136]. The positively charged Dab residues of the polypeptide provide a means of electrostatic attraction with the negatively charged phosphate headgroups of lipid A [137]. This binding induces competitively the displacement of divalent cations (Ca++ and Mg++) from the phosphate groups of membrane lipids [138,139] (Figure 3). Colistin molecules insert the hydrophobic N-terminal fatty acyl chain segment into the outer membranes leading to local destabilized areas of the membrane [140,141]. This induced disruption of the outer cell membrane causes an increase in the permeability of the cell envelop, followed by leakage of cell content, and eventually, cell lysis [141, 142] (Figure 3). However, it is important to note that recent studies showed that colistin might exert its antibacterial effect through an alternative route of action [143]. Thus, the exact mechanism of action of colistin needs further investigations.
Mechanisms of Resistance
The prevalence of resistance to colistin is relatively rare and the data is very scarce. This probably reflects the uncommon use of the drug worldwide. However, the past years have witnessed a growth in the incidence rate of colistin resistance following the failure of MDR Gram-negative bacteria to respond to all currently available antibacterial therapeutics [144]. In fact, Colistin-resistant isolates have been recently identified in Gram-negative bacteria species such as A. baumannii, K. pneumoniae and P. aeruginosa, and many other strains to be discussed in following sections [145].
Natural or Intrinsic Resistance: Until the discovery of plasmid mediated resistance, Colistin was only known for its chromosomally mediated, inherited resistance via vertical transmission. There is a number of Gram-negative bacteria that are naturally resistant to colistin; these mainly include the following: Neisseria spp., Moraxella catarrhalis, Helicobacter pylori, Proteus mirabilis, Serratia marcescens, Morganella morganii, Chromobacterium and Brucella species [146,147].
Acquired Resistance: The general mechanism by which bacteria acquires resistance against colistin is currently under extensive research. The sudden spread of resistance is of a primary global concern and is believed to involve upregulation of a number of regulatory systems [148]. We previously described that the critical and essential step in the mechanism of action of the drug is the electrostatic interactions that take place between positively charged residues on colistin and the negatively charged phosphate group of lipid A on LPS [149,150]. In fact, resistant species were able to alter this type of interactions through lipid A modification [151]. This reduces the total negative net charge of the outer membrane and therefore weakens the attraction between the drug and its target [152]. Recently, the most common mechanism of resistance to colistin is the acquisition of plasmid-mediated resistant gene through horizontal gene transfer, mainly conjugation [153, 154].
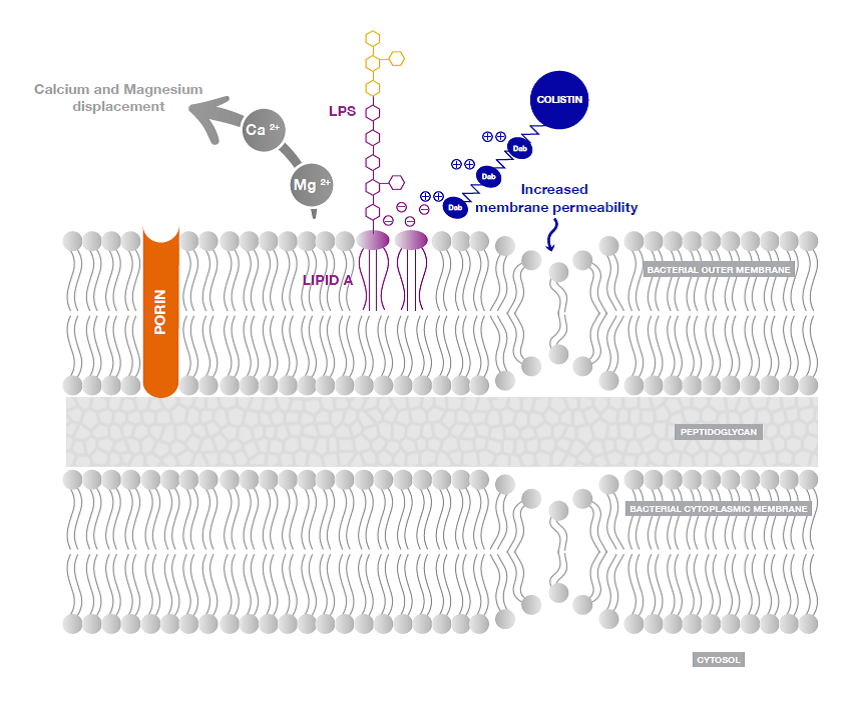
Figure 3: Action of colistin on Gram-negative bacterial membrane [2].
Cationic colistin binds with anionic Lipopolysaccharide (LPS) molecules by displacing calcium and magnesium from the outer cell membrane of Gram-negative bacteria, leading to permeability changes in the cell envelope and leakage of cell contents. By binding to the Lipid A portion of LPS, colistin also has an anti-endotoxin activity.
The Colistin Resistant Gene
Overview: Until recently, polymyxin resistance has been always reported via chromosomal mutations but has never shown to involve horizontal gene transfer. A significant increase in colistin was observed in commensal E.coli from food animals in China during a routine surveillance project on antimicrobial resistance. Analysis of an E. coli strain isolated from a pig, possessing colistin resistance, showed the emergence of the first plasmid-mediated polymyxin resistance mechanism, designated mcr-1, in Enterobacteriaceae.
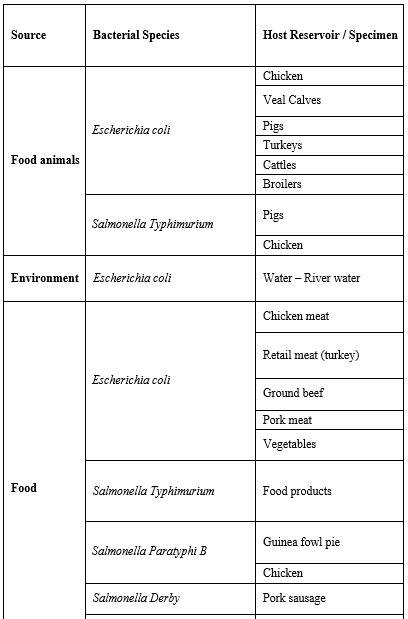
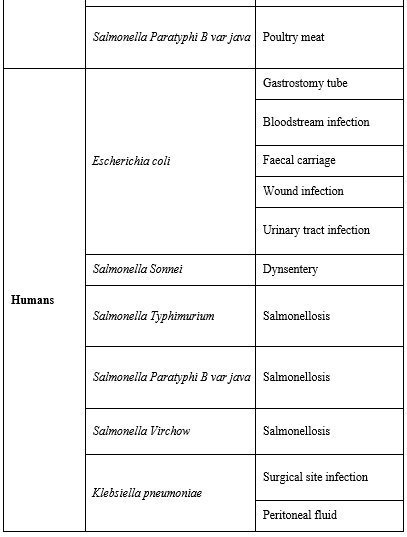
Bacterial Species Carrying mcr-1: Interestingly, mcr-1 was shown to have a very high in-vitro transfer rate between E. coli strains and is capable as well to transfer into epidemic strains of Enterobacteriaceae, such as Klebsiella pneumonia, Pseudomonas aeruginosa, and many others listed in (Table 1) [155].
Animal to Human Transmission: Based on the fact that mcr-1 gene predominates and was first described in animals; it was suggested that plasmid mediated colistin resistance moved from animals to humans [155]. In addition, animals are reported to consume the largest volume of colistin antibiotic [156]. Moreover, colistin resistance in bacterial isolates from animal feeds confirmed the contribution of antibiotic consumption in animals to increased resistance levels in humans [157].
Table 1: Bacterial species reported carrying mcr-1 gene and their host reservoir [16].
Mechanism of Resistance and Catalytic Structure of mcr-1
As mentioned previously, modification of the LPS component of the outer membrane is the most common mechanism of acquired resistance. Resistance takes place when the 1’ and 4’ phosphate groups of lipid A are modified resulting in neutralizing the negative charge and reducing the binding of the positively charged colistin [158,159]. This mechanism is associated with gene mutations that result in expression of transferase enzymes that modify the LPS [158,159]. The phosphate groups are modified by the phosphoethanolamine transferase enzymes that add phosphoethanolamine (PEA) or by aminoarabinose transferase enzymes (ArnT) that add 4-aminoarabinose [160, 161]. ArnT belongs to the GT-C family of glycosyltransferases. It is a membrane protein having a periplasmic domain. PEA transferases have a periplasmic catalytic domain in addition to a membrane spanning one [162, 163]. mcr-1 gene, a plasmid-mediated resistance gene in Escherichia coli isolates from humans and animals, was identified in China [155]. mcr-1 confers resistance by modification of the colistin target. As shown in Figure 3, it catalyzes the transfer of phosphoethanolamine (PEA) onto the glucosamine saccharide of lipid A in the bacterial outer membrane [155]. The resulting reduced net charge of the outer membrane disrupts colistin binding [164]. The structures of the catalytic domains of Campylobacter jejuni PEA transferase “EptC” and Neisseria meningitidis PEA transferase “LptA” as well as an ArnT transferase were previously identified and described [160, 161]. LptA and EptC PEA transferases have catalytic domains similar in structure and are members of the sulfatase group. The MCR-1 enzyme was found to be 41 % identical to the PEA transferase LptA and 40 % to EptC [160] with both their sequence comparisons suggesting conservation of the active-site residues. Palzkill and colleagues resolved the X-ray crystal structure of the soluble periplasmic catalytic domain of the MCR-1 [165]. They identified a globular protein with an overall hemispherical shape having a central β-sheet composed of seven β-strands sandwiched between α-helical structures of a cMCR [165]. Moreover, they detected the presence of a phosphate group attaching covalently to the side-chain hydroxyl oxygen, which results from phosphorylation of Thr285 [165]. In addition, three zinc ions were found surrounding the phosphothreonine. The presence of zinc binding sites and phosphorylated threonine along with previous studies of PEA transferases, confirmed that region of the catalytic domain is the active site of MCR-1 [163, 166]. Based on amino acid sequence homology, LptA from Neisseria meningitidis and PEA transferases EptC from Campylobacter jejuni are found to be the closest in structure to MCR-1, thus being noted to be homologs [160,163,166]. In summary, the structure of the catalytic domain of MCR-1 reveals conservation of structure, particularly in the active site, with other PEA transferases [163, 166]. Philip Hinchliffe and his colleagues again confirmed this work. They presented crystal structures showing the MCR-1 periplasmic, catalytic domain to be a zinc metalloprotein with an alkaline phosphatase/sulphatase fold containing three disulphide bonds [167]. To understand the impact of MCR-1 activity across the range of Gram-negative bacteria and to identify alternative means to overcome its activity, they investigated the molecular basis of MCR-1. Mutation of residues implicated in zinc or phosphoethanolamine binding, or catalytic activity, resulted in the recovery of colistin susceptibility of recombinant E. coli [167,168]. These results show the importance of PEA transferases as interesting drug target for they are in most Gram-negative bacteria and have important role in mechanisms of bacterial lipopolysaccharide modification [166]. In addition, the use of inhibitors of MCR-1 may restore polymyxin susceptibility.
mcr-2: The Other Colistin Resistant Gene
Identification: After the reporting of a colistin-resistant mcr-1 gene in China [169], S. Malhotra-Kumar and colleagues screened 105 colistin resistant E. coli extracted from 52 calves and 53 piglets from different regions of Belgium [170]. 13 isolates revealed the presence of mcr-1 and the others were preserved for later studies for the cause of resistance to colistin [170, 171].
In another study performed by the same group, 10 porcine and bovine colistin-resistant Escherichia coli isolates were randomly selected and their plasmid DNA was amplified. The results showed three of the ten isolates harboring the presence of a gene coding for phosphoethanolamine transferase enzyme, however, it was not mcr-1. The novel plasmid-mediated colistin resistance-conferring gene, was named mcr-2 [172].
Structural Analyses of the MCR-2 Protein: S. Malhotra-Kumar and his team studied the similarities of MCR-1 and MCR-2 and showed that both proteins had 80.65% identity [172]. Moreover, mcr-2 gene was found 1,617 bp long, shorter than mcr-1 by nine bases, with 76.75% identity to mcr-1 [172]. It was predicted to have two domains, with domain 1 as a transporter domain that would be embedded in the cell membrane and responsible for transporting different molecules, and domain 2 as a transferase domain for the enzyme [172]. S. Malhotra-Kumar and colleagues conducted Phylogenetic analysis - the study of the evolutionary history and relationships among species – and revealed that mcr-2 might have originated from Moraxella catarrhalis [172]. Phylogenetic analysis between mcr-1 and mcr-2 provided strong evidence that the latter was distinct from mcr-1.
Predicted Tertiary Structure: Moreover, the same study predicted that MCR-2 protein has two domains: (i) domain 1 as a transporter, with residues ranging between 1 to 229; and (ii) domain 2 as a transferase domain, with residues ranging between 230 to 538 [172]. The best template for domain 1 was 4HE8. 4HE8 is a secondary membrane transport protein that transfers solutes across membranes [173]. The best-fit template for domain 2 was 4kav, a lipo-oligosaccharide phosphoethanolamine transferase A from Neisseria meningitidis. 4kav was previously detected to be the best fit template for MCR-1 [169].
Discussion
Antimicrobial resistance is the ability of infectious organisms to survive the drugs that are designed to kill them, or inhibit their growth, and therefore render the infection treatable [169]. The rapid diffusion and dissemination of Antibiotic resistance and the emergence of multidrug resistant bacteria represent a threat of great importance to global health [174]. While the phenomenon can occur naturally through bacterial adaptation to environmental conditions, development of resistant genes from natural selection aspect showed low incidence compared to the high incidence level of acquired resistance via transfer of plasmids containing resistant genes among closely as well as distantly related species [175]. Carbapenem and Colistin serve as last-resort antibiotics against infections caused by MDR Gram-negative bacteria [176]. Production of B-lactamases is one of the most common mechanisms used by Gram-negative bacteria for resistance against beta-lactam antibiotics such as carbapenem [177]. Over the years, various beta-lactamases and carbapenemases were identified in Gram-negative bacteria with several species displaying co-resistance to most available drugs, thus limiting medication options [178]. The novel carbapenemase New Delhi metallo-b-lactamases (NDM-1), encoded by the blaNDM-1 gene, has been reported in many pathogenic bacterial species, especially members of Enterobacteriacea [179]. In infections due to NDM-positive bacteria, antimicrobial choice is limited to polymyxins mainly, especially polymyxin E, Colistin. For years, clinical use of colistin was shown to be effective against NDM-producing bacteria; Polymyxins were increasingly used as last resort for infections caused by these species harboring blaNDM resistant gene [180, 181]. Unfortunately, discovery of transmissible colistin resistance in China and subsequent identification of plasmid-borne gene mcr-1 was reported in animal, food, and clinical samples, worldwide [182,183]. Moreover, following reports of mcr-1 detection, a team of researchers in Belgium reported discovery of a new gene with the ability to confer colistin resistance; phylogenic analysis showed that this gene was distinct from mcr-1 and was termed mcr-2 [184]. Further studies indicated that mcr-2 gene might be able to spread more rapidly than mcr-1; prevalence data showed mcr-2 has a higher transfer frequency. Most recently, mcr-1 gene was identified in two clinical carbapenem-susceptible E.coli strains in United States. The two strains were isolated from urine cultures obtained in May 2015 and April 2016 [185, 186]. In October 2016, two colistin-resistant E. coli isolates were reported in Venezuela; the report represents the first detection of mcr-1 in the country. Moreover, one of the mcr-1-positive E. coli strains showed to harbor blaNDM-1, revealing the coexistence of the two genes within the same isolate [187]. Therefore, with the emergence of species harboring genes resistant to both carbapenem and colistin creates a highly drug-resistant member that is potentially untreatable and poses a serious threat to public health worldwide [188, 189]. The coexistence of mcr-1 with other resistance genes with high potential of spread is of great concern. Nevertheless, in a recent study published in February 2017, Zhu and colleagues investigated the effect of a combination therapy of colistin and another class of antibiotics amikacin against E. coli strain expressing mcr-1 and blaNDM-5 – variant of blaNDM-1 – resistant genes. In vitro assays and in vivo analyses in mice revealed a better response to combined therapy compared to monotherapy: use of colistin and amikacin separately. In fact, augmented susceptibility was recorded in strains treated with combination therapy indicating that it may be a promising option [190]. Recent advances in genome sequencing and gene editing tools allowed researchers to identify genetic changes corresponding to antibiotic resistance and carry out genetic manipulations in the aim of restoring antimicrobial activity [191, 192]. In one study published earlier this year - January 2017- in the Journal of Antimicrobial Chemotherapy, Geller and his colleagues at the Oregon State University (OSU) developed a new molecule with the ability to reverse resistance depicted by the presence and expression of blaNDM-1 gene [193]. The molecule is a type of an oligomer, which has the ability to bind complementary to mRNA sequences and exert its effect through inhibition of translation [194]. In this study, the so-called Peptide Phosphorodiamidate Morpholino Oligomers (PPMOs) were described to inhibit the expression of NDM-1. In vitro analyses revealed the great potential of the developed molecule in restoring susceptibility to meropenem – class of carbapenem – by making bacteria sensitive to the drug it was resistant to. The study combined meropenem with PPMO and directed it against three different types of NDM positive bacteria; in all cases, PPMO restored meropenem activity and resistance was inhibited [194]. PPMO represents a new hope in the absence of rapid development of antibiotics that would show effectiveness against all resistant species. However, the prevalence of antibiotic resistance genes and its widespread progression rate poses serious pressure on the need for immediate global actions in the aim of fighting against antimicrobial resistance.
Conclusion
In this review, we have collected available data resources to address the knowledge regarding the blaNDM and mcr-1 and mcr-2 genes resistant to last-resort antibiotics carbapenem and colistin respectively. Resistant genes do not recognize geographical and ecological borders; the widespread of these genes is increasing at a rate that exceeds development of potential drugs effective against corresponding infections. Consequences include failure to successful treatment and therefore prolonged illness and increased mortality rate. A collaborative network to support surveillance of antimicrobial resistance, development of low-cost diagnostic tools along with other medicinal interventions are required for improved assessment and containment of the situation. Moreover, a global action plan to limit inappropriate use of these drugs in food, agriculture, and animal farming to restrict the further progression and transfer among different species.
List of Abbreviations
Anti-Microbial Resistance (AMR), Extended-Spectrum Beta-Lactamase (ESBL), AR: Antibiotic Resistance (AR), World Health Organization (WHO), Antimicrobial resistance (AMR), Antibiotic resistance genes (ARG), Aminoarabinose transferase enzymes (ArnT), New Delhi metallo-beta-lactamase (blaNDM), Colistimethate sodium (CMS), Esherishia coli (E. coli), Extended spectrum β-lactamases (ESBLs), Gastrointestinal tract (GIT), Horizontal Gene Transfer (HGT), Integrative conjugative elements (ICEs), Lipooligosaccharide phosphoethanolamine transferase (LptA), Lipopolysaccharides (LPS), Mobile colistin resistance-1 (mcr-1), Mobile colistin resistance-2 (mcr-2), Metallo-beta-lactamses (MBLs), Mobile genetic elements (MGEs), Multidrug resistant (MDR), N-acetylmuramic acid (NAM), N- acetylglucosamine (NAG), Oregon State University (OSU), Phosphoethanolamine (PEA), Penicillin binding proteins (PBP), Peptidoglycan (PPG), Peptide phosphorodiamidate morpholino oligomers (PPMOs), Polymyxin B (PMB), Polymerase Chain Reaction (PCR). Threonine (Thr), UDP-N-acetylmuramyl (UDP-MurNAc), UDP-N-acetylglucosamine (UDP-GlcNAc), Urinary tract infection (UTI), Verona integron-encoded metallo-β-lactamase (VIM), Vertical Gene Transfer (VGT).
Declarations
Competing interests
Authors declare having no competing interests.
Grant information
The authors received no specific funding for this work.
Authors’ contributions
S.Sakr and B. Hamam designed and supervised the study. C. Mansour, K. Mansour and J. Kobeissi drafted the manuscript, L. Saliba edited the pictures, R. Bou Raad and I. Sheet edited the manuscript. All authors have approved the final version of manuscript.
References
-
-
- Silhavy TJ, Kahne D and Walker S. The Bacterial Cell Envelope. CSHPB. 2010;2(5).
- Wang S, Arellano-Santoyo H, Combs PA and Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. PNAS U S A. 2010;107(20):9182‐5.
- Turner RD, Hurd AF, Cadby A, Hobbs JK and Foster SJ. Cell Wall Elongation Mode in Gram-negative Bacteria is Determined by Peptidoglycan Architecture. NC. 2013;4:1496.
- Miller SI. Antibiotic Resistance and Regulation of the Gram-Negative Bacterial Outer Membrane Barrier by Host Innate Immune Molecules. mBio. 2016;7(5):e01541-16.
- Moyes RB, Reynolds J and Breakwell DP. Differential Staining of Bacteria: Gram Stain. CPM. 2009;15:3.
- Matsuura, M. Structural modifications of bacterial lipopolysaccharide that facilitate gram-negative bacteria evasion of host innate immunity. FI. 2013;4:109.
- Bertani B and Ruiz N. Function and biogenesis of lipopolysaccharides. ESP. 2018;8(1).
- Clayton L Thomas. Taber's Cyclopedic Medical Dictionary (17th ed.). F. A. Davis Co. 1993. ISBN 0-8036-8313-8.
- Hopkins SJ. Drugs and Pharmacology for Nurses (12th ed.). Churchill Livingstone.1997. ISBN 0-443-05249-2.
- Ventola CL. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. P&T. 2015;40(4):277-83.
- Van Elsas JD and Bailey MJ. The ecology of transfer of mobile genetic elements. FEMS ME. 2002; 42:187-197.
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a New Metallo-Beta-Lactamase Gene, bla(NDM-1), and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella Pneumoniae Sequence Type 14 From India. AAC. 2009;53(12):5046-54.
- Govindaswamy A, Bajpai V, Khurana S, Batra P, Mathur P and Malhotra R. Prevalence and Characterization of Carbapenemase-producing Escherichia coli from a Tertiary Care Hospital in India. JGID. 2019;11(3):123-4.
- Hasman H, Hammerum A, Hansen F, Hendriksen R, Olesen B, Agersø Y, et al. Detection of mcr-1 Encoding Plasmid-Mediated Colistin-Resistant Escherichia Coli Isolates From Human Bloodstream Infection and Imported Chicken Meat, Denmark 2015. Euro Surveill. 2015;20(49).
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. ES. 2016;21(27):30280.
- Woo P, To A, Lau S and Yuen K. Facilitation of Horizontal Transfer of Antimicrobial Resistance by Transformation of Antibiotic-Induced Cell-Wall-Deficient Bacteria. MH. 2003;61(4):503-8.
- Davies J and Davies D. Origins and Evolution of Antibiotic Resistance. ASM. 2010;74(3):417-33.
- Karlsson E, Kwiatkowski D and Sabeti P. Natural Selection and Infectious Diseases in Human Populations. 2014;15(6):379-93.
- Forsberg K, Patel S, Gibson M, Lauber C. Knight R, Fierer N, et al. Bacterial Phylogeny Structures Soil Resistomes Across Habitats. Nature. 2014;509(7502):612-6.
- Baltrus D. Exploring the Costs of Horizontal Gene Transfer. TEE. 2013;28(8):489-95.
- Norman A, Hansen L and Sorensen S. Conjugative Plasmids: Vessels of the Communal Gene Pool. PTRSLBBS. 2009;364(1527):2275-89.
- Arutyunov D and Frost L. F Conjugation: Back to the Beginning. Plasmid. 2013;70(1):18-32
- Norman A, Hansen L and Sorensen S. Conjugative Plasmids: Vessels of the Communal Gene Pool. PTRSLBBS. 2009;364(1527):2275-89.
- Sorensen SJ, Bailey M, Hansen LH, Kroer N and Wuertz S. Studying Plasmid Horizontal Transfer in Situ: A Critical Review. NRM. 2005;3(9):700-10.
- Barlow M. What Antimicrobial Resistance Has Taught Us About Horizontal Gene Transfer. MMB. 2009;532:397-411.
- Nojiri H, Shintani M and Omori T. Divergence of Mobile Genetic Elements Involved in the Distribution of Xenobiotic-Catabolic Capacity. AMB. 2004;64(2):154-74.
- Weinert LA, Welch J, and Jiggins FM. Conjugation Genes Are Common Throughout the Genus Rickettsia and Are Transmitted Horizontally. PBS. 2006;276(1673):3619-27.
- Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of Plasmids. 2010;74(3):434-52.
- Carattoli A. Plasmids and the Spread of Resistance. IJMM. 2010;303(6-7):298-304.
- Wiedenbeck J, Cohan FM. Origins of Bacterial Diversity Through Horizontal Genetic Transfer and Adaptation to New Ecological Niches. FEMS MR. 2011;35(5):957-76.
- Treangen TJ and Rocha EPC. Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in Prokaryotes. PLoS G. 2011;7(1):e1001284.
- Martinez JL. Antibiotics and Antibiotic Resistance Genes in Natural Environments. Science. 2008;321(5887):365-7.
- San Millan A, Heilbron K and MacLean RC. Positive Epistasis Between Co-Infecting Plasmids Promotes Plasmid Survival in Bacterial Populations. ISME J. 2014;8(3):601-12.
- Wozniak RA and Waldor MK. Integrative and Conjugative Elements: Mosaic Mobile Genetic Elements Enabling Dynamic Lateral Gene Flow. NRM. 2010;8(8):552-63.
- Steffee CH, Alexander Fleming and Penicillin. The Chance of a Lifetime? N C M J. 1992; 53(6):308-10.
- Fleming A. On the Antibacterial Action of Cultures of a Penicillium, With Special Reference to Their Use in the Isolation of B. Influenzae. 1929. BWHO. 2001;79(8):780-90.
- Hare R. New Light on the History of Penicillin. MH. 1982;26(1):1-24.
- Queener SF. History and origins of beta-lactam antibiotics. In: Queener SF, Webber JA, Queener SW, editors. Beta-Lactam Antibiotics for Clinical Use. New York: Marcel Dekker, Inc. 1986;3-16.
- Florey HW, Heatley NG, Jennings MA, Sanders AG, Abraham EP and Florey ME. Historical introduction Antibiotics. Oxford University Press. 1949;2:768.
- Chain E. The early years of the penicillin discovery. Trends Pharmacol Sci J. 1979;1:6-11.
- Bush K. The Coming Age of Antibiotics: Discovery and Therapeutic Value. ANYAS. 2010;1213:1-4.
- Mahajan GB and Balachandran L. Antibacterial Agents from Actinomycetes- A Review. FB. 2012;4:240-53.
- Torres J, Villegas M and Quinn J. Current Concepts in Antibiotic-Resistant Gram-Negative Bacteria. ERAIT. 2007;5(5):833-43.
- Papp-Wallace KM, Endimiani A, Taracila MA and Bonomo RA. Carbapenems: Past, Present, and Future. AAC. 2011;55(11):4943-60.
- Rogers HJ, Perkins HR and Ward JB. Microbial cell walls and membranes. Chapman and Hall, New York, N.Y. 1980.
- Typas A, Banzhaf M and Gross CA. Vollmer W. From the Regulation of Peptidoglycan Synthesis to Bacterial Growth and Morphology. NRM. 2011;10(2):123-36.
- Lovering AL, Safadi SS and Strynadka NCJ. Structural Perspective of Peptidoglycan Biosynthesis and Assembly. ARB. 2012;81:451-78.
- Kuru E, Hughes H, Brown P, Hall E, Tekkam S, Cava F, et al. In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D-Amino Acids. ACIEE. 2012;51(50):12519-23.
- Atilano M, Pereira P, Yates J, Reed P, Veiga H, Pinho M, et al. Teichoic Acids Are Temporal and Spatial Regulators of Peptidoglycan Cross-Linking in Staphylococcus Aureus. PNAS USA. 2010; 107(44):18991-6.
- Shi Q, Meroueh SO, Fisher JF and Mobashery S. A Computational Evaluation of the Mechanism of Penicillin-Binding Protein Catalyzed Cross-Linking of the Bacterial Cell Wall. JACS. 2011; 133(14):5274-83.
- Buynak JD. Cutting and Stitching: The Cross-Linking of Peptidoglycan in the Assembly of the Bacterial Cell Wall. ACS CB. 2007;2(9):602-5.
- Goffin C and Ghuysen JM. Multimodular Penicillin-Binding Proteins: An Enigmatic Family of Orthologs and Paralogs. MMBR. 1998;62(4):1079-93.
- Mahajan GB and Balachandran L. Antibacterial Agents from Actinomycetes- A Review. FB. 2012; 4:240-53.
- Walsh C and Walsh C. Antibiotics: actions, origins, resistance. ASM. Washington, D.C. 2003.
- Abraham EP and Chain E. An Enzyme from Bacteria Able to Destroy Penicillin. 1940. RID. 1988; 10(4):677-8.
- Hakenbeck R, Bruckner R, Denapaite D and Maurer P. Molecular Mechanisms of Β-Lactam Resistance in Streptococcus Pneumoniae. FM. 2012;7(3):395-410.
- Misra R and Bavro VN. Assembly and Transport Mechanism of Tripartite Drug Efflux Systems. BBA. 2009;1794(5):817-25.
- Wilke MS, Lovering AL and Strynadka NCJ. Beta-lactam Antibiotic Resistance: A Current Structural Perspective. COM. 2005;8(5):525-33.
- Fuda C, Surorov M, Vakulenko SB and Mobashery S. The Basis for Resistance to Β-Lactam Antibiotics by Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus Aureus. JBC. 2004;279(39):40802-6.
- Smith CA, Frase H, Toth M, Kumarasiri M, Wiafe K, Munoz J, et al. Structural Basis for Progression Toward the Carbapenemase Activity in the GES Family of Β-Lactamases. JACS. 2012;134(48):19512-5.
- Lee JH, Bae IK and Lee SH. New Definitions of Extended-Spectrum Β-Lactamase Conferring Worldwide Emerging Antibiotic Resistance. MRR. 2012;32(1):216-32.
- Biehl LM, Schmidt-Hieber M, Liss B, Cornely OA and Vehreschild M. Colonization and Infection with Extended Spectrum Beta-Lactamase Producing Enterobacteriaceae in High-Risk Patients- Review of the Literature from a Clinical Perspective. CRM. 2016;42(1):1-16.
- Chouchani C, Marrakchi R and El Salabi A. Evolution of Beta-Lactams Resistance in Gram-negative Bacteria in Tunisia. CRM. 2011;37(3):167-77.
- Ambler RP, Coulson FW, Frere J-M, Ghuysen J-M, Joris B, Forsman M, et al. A Standard Numbering Scheme for tClass A Β-Lactamases. BJ. 1991;276:269-70.
- Bush K and Jacoby GA. Updated Functional Classification of Beta-Lactamases. AAC. 2010;54(3):969-76.
- Ghuysen J-M. Serine β-Lactamases and Penicillin-Binding Proteins. ARM. 1991;45:37-67.
- Mitchell JB. The Relationship Between the Sequence Identities of Alpha Helical Proteins in the Pdb and The Molecular Similarities of Their Ligands. JCICS. 2001;41(6):1617-22.
- Crowder MW, Spencer J and Vila AJ. Metallo-β-lactmases: Novel Weaponry for Antibiotic Resistance in Bacteria. ACR. 2006;39(10):721-8.
- Wang Z, Fast W, Valentine AM and Benkovic SJ. Metallo-β-lactamase: Structure and Mechanism. COCB. 1999;3(5):614-22.
- Cole M. ‘Beta-lactams’ as Beta-Lactamase Inhibitors. PTRSLBBS. 1980;289(1036):207-23.
- Richet HM, Mohammed J, McDonald LC and Jarvis WR. Building Communication Networks: International Network for The Study and Prevention of Emerging Antimicrobial Resistance. EID. 2001;7(2):319-22.
- Jeon JH, Lee JH, Lee JJ, Park KS, Karim A, et al. Structural Basis for Carbapenem-Hydrolyzing Mechanisms of Carbapenemases Conferring Antibiotic Resistance. IJMS. 2015;16(5):9654-92.
- Patel G and Bonomo RA. Status Report on Carbapenemases: Challenges and Prospects. ERAIT. Ther. 2011;9(5):555-70.
- Nordmann P, Dortet L and Poirel L. Carbapenem Resistance in Enterobacteriaceae: Here is the Storm! TMM. 2012;18(5):263-72.
- Papp-Wallace KM, Endimiani A, Taracila MA and Bonomo RA. Carbapenems: Past, Present, and Future. AAC. 2011;55(11):4943-60.
- Byeong CJ, Sang HL. Structural Basis for Carbapenem-Hydrolyzing Mechanisms of Carbapenemases Conferring Antibiotic Resistance. IJMS. 2015;16(5):9654-92.
- Tremblay LW, Fan F and Blanchard JS. Biochemical and Structural Characterization of Mycobacterium Tuberculosis β-Lactamase with the Carbapenems Ertapenem and Doripenem. Biochemistry. 2010;49(17):3766-73.
- Kuwabara S and Abraham EP. Some Properties of Two Extracellular B-Lactamases from Bacillus Cereus 569/H. BJ. 1967;103(3):27C-30C.
- Lim HM, Pene JJ and Shaw R. Cloning, Nucleotide Sequence, and Expression of the Bacillus Cereus 5/B/6 b-Lactamase II Structural Gene. JB. 1988;170(6):2873-8.
- Iaconis JP and Sanders CC. Purification and Characterization of Inducible Beta-Lactamases in Aeromonas Spp. AAC. 1990;34(1):44-51.
- Yang Y and Bush K. Biochemical Characterization of the Carbapenem-Hydrolyzing Beta-Lactamase AsbM1 From Aeromonas Sobria AER 14M: A Member of a Novel Subgroup of Metallo-Beta-Lactamases. FEMS M L. 1996;137(2-3):193-200.
- Saino Y, Kobayashi F, Inoue M and Mitsuhashi. Purification and Properties of Inducible Penicillin Beta-Lactamase Isolated from Pseudomonas Maltophilia. AAC. 1982;22(4):564-70.
- Nordmann P, Cuzon G and Naas T. The Real Threat of Klebsiella Pneumoniae Carbapenemase-Producing Bacteria. LID. 2009;9(4):228-36.
- Nordmann P, Dortet L and Poirel L. Carbapenem Resistance in Enterobacteriaceae: Here is the Storm! TMM. 2012;18(5):263-72.
- Nordmann P and Poirel L. The Difficult-To-Control Spread of Carbapenemase Producers Among Enterobacteriaceae Worldwide. CMI. 2014;20(9):821-30.
- Laraki N, Galleni M, Thamm I, Riccio ML, Amicosante G, et al. Structure of In31, a blaIMP-containing Pseudomonas Aeruginosa Integron Phyletically Related to In5, Which Carries an Unusual Array of Gene Cassettes. AAC. 1999;43(4):890-901.
- Borra PS, Leiros HKRA, Spencer J, Leiros I, Walsh TR, Sundsfjord A, et al. Structural and Computational Investigations of VIM-7: Insights Into the Substrate Specificity of Vim Metallo- Β-Lactamases. JMB. 2011;411(1):174-89.
- Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, et al. Cloning and characterization of blaVIM, a New Integron-Borne Metallo-Beta-Lactamase Gene From a Pseudomonas Aeruginosa Clinical Isolate. AAC. 1999;43(7):1584-90.
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a New Metallo-Beta-Lactamase Gene, bla(NDM-1), and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella Pneumoniae Sequence Type 14 From India. AAC. 2009; 53(12):5046-54.
- Nordmann P, Poirel L and Walsh TR. Livermore DM. The Emerging NDM Carbapenemases. TM. 2011;19(12):588-95.
- Nordmann P, Poirel L, Toleman MA and Walsh TR. Does Broad-spectrum Beta-Lactam Resistance Due to NDM-1 Herald the End of The Antibiotic Era for Treatment of Infections Caused by Gram-negative Bacteria? JAC. 2011;66(4):689-92.
- Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC and Lee SH. Global Dissemination of Carbapenemase-Producing Klebsiella Pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. FM. 2016;7:895.
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. LID. 2010;10(9):597-602.
- Wilson ME and Chen LH. NDM-1 and the role of travel in its dissemination. CIDR. 2012;14(3):213-26.
- Cornaglia G, Giamarellou H and Rossolini GM. Metallo-Beta-Lactamases: A Last Frontier for Beta-Lactams? LID. 2011;11(5):381-93.
- Khan AU and Nordmann P. Spread of Carbapenemase NDM-1 Producers: The Situation in India and What May be Proposed. SJID. 2012;44(7):531-5.
- Livermore DM, Walsh TR, Toleman M and Woodford N. Balkan NDM-1: Escape or Transplant? LID. 2011;11(3):164.
- Nordmann P, Poirel, L and Walsh TR. Livermore DM. The Emerging NDM Carbapenemases. TM. 2011;19(12):588-95.
- Johnson AP and Woodford N. Global Spread of Antibiotic Resistance: The Example of New Delhi Metallo-Beta-Lactamase (NDM)-Mediated Carbapenem Resistance. JMM. 2013;62:499-513.
- Patel G and Bonomo RA. "Stormy Waters Ahead": Global Emergence of Carbapenemases. FM. 2013;4:48.
- Chen LH and Wilson ME. The Role of the Traveler in Emerging Infections and Magnitude of Travel. MCNA. 2008;92(6):1409-32.
- Reed CM. Medical tourism. MCNA. 2008;92(6):1433-46.
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of A New Antibiotic Resistance Mechanism in India, Pakistan, and The UK: A Molecular. LID. 2010; 10(9):597-602.
- Flateau C, Janvier F, Delacour H, Males S, Ficko C, Andriamanantena D, et al. Recurrent Pyelonephritis Due to NDM-1 Metallo-Beta-Lactamase Producing Pseudomonas Aeruginosa in A Patient Returning From Serbia, France. ES. 2012:17.
- Pitout JD, Nordmann P and Poirel L. Carbapenemase-Producing Klebsiella pneumoniae, a Key pathogen set for global nosocomial dominance. AAC. 2015;59:5873-84.
- Nordmann P and Poirel L. The Difficult-To-Control Spread of Carbapenemase Producers Among Enterobacteriaceae Worldwide. CMI. 2014;20(9):821-30
- Kazi M, Drego L, Nikam C, Ajbani K, Soman R, Shetty A, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. EJCMD. 2015;34:467-72.
- Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, et al. New Delhi Metallo-b-lactamase around the world: an eReview using Google Maps. Euro. Surveill. 2014;19:20809.
- Peirano G, Ahmed-Bentley J, Fuller J, Rubin JE and Pitout JD. Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. JCM. 2014;52:1575-81.
- Sprignani J, Magistro A, Dal Peraro M, Vila A, Carloni P and Pierattelli R. On the Active Site of Mononuclear B1 Metallo β-Lactamases: A Computational Study. JCAMD. 2012;26:425-35
- Suárez D, Brothers EN and Merz KM Jr. Insights Into the Structure and Dynamics of the Dinuclear Zinc Beta-Lactamase Site From Bacteroides Fragilis. Biochemistry. 2002;41(21):6615‐30.
- Jeon JH, Lee JH, Lee JJ, Park KS, Karim A, Lee CR, et al. Structural Basis for Carbapenem-Hydrolyzing Mechanisms of Carbapenemases Conferring Antibiotic Resistance. IJMS. 2015;16(5):9654-92.
- Jeon JH, Lee JH, Lee JJ, Park KS, Karim A, Lee CR, et al. Structural Basis for Carbapenem-Hydrolyzing Mechanisms of Carbapenemases Conferring Antibiotic Resistance. IJMS. 2015;16(5):9654-92.
- Hinchliffe P, Tanner C, Krismanich A, Labbé G, Goodfellow V, Marrone L, et al. Structural and Kinetic Studies of the Potent Inhibition of Metallo-β-lactamases by 6-Phosphonomethylpyridine-2-carboxylates. Biochemistry. 57(12):1880-92.
- Mojica M, Bonomo R and Fast W. B1-Metallo-β-Lactamases: Where Do We Stand? CDT. 2016;17(9):1029-50.
- Li J, Nation RL, Milne RW, Turnidge JD and Coulthard K. Evaluation of colistin as an agent against multi- resistant Gram-negative bacteria. Int J Antimicrobial Agents. 2005;25:11-25.
- Falagas ME and Kasiakou SK. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. CID. 2005;40:1333-41.
- Orwa JA, Govaerts C, Busson R, Roets E, Van Schepdael A and Hoogmartens J. Isolation and structural characterization of colistin components. JA, 2001;54(7):595-9.
- Velkov T, Roberts KD, Nation RL, Ompson PE and Li J. Pharmacology of polymyxins: new insights into an “old” class of antibiotics. FM. 2013;8(6):711-24.
- Nation RL and Li J. Colistin in the 21st century. COID. 2009;22:535-43.
- Koch-Weser J, Sidel VW, Federman EB, Kanarek P, Finer DC and Eaton AE. Adverse effects of sodium colistimethate. Manifestations and specific reaction rates during 317 courses of therapy. AIN. 1970;72:857-68.
- Boucher HW, Talbot GH and Bradley JS. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. CID. 2009; 48:1-12.
- Li J, Nation RL and Turnidge JD. Colistin: the re-emerging antibiotic for multidrug resistant Gram- negative bacterial infection. Lancet. 2006;9(6):589- 601.
- Choi SK, Park SY and Kim R. Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. JB. 2009;191(10):3350-8.
- Orwa JA, Govaerts C, Busson R, Roets E, Van Schepdael A and Hoog-martens J. Isolation and structural characterization of colistin components. JA. 2001;54:595-9.
- Karvanen M. Optimization of Colistin Dosage in the Treatment of Multiresistant Gram-negative Infections. AUU. 56.
- Okimura K, Ohki K, Sato Y, Ohnishi K and Sakura N. Semi-synthesis of polymyxin B (2–10) and colistin (2–10) analogs employing the Trichloroethoxycarbonyl (Troc) group for side chain protection of α,γ-diaminobutyric acid residues. CPB. 2007;55(12):1724-30.
- Mohamed AF, I Karaiskos, D Plachouras, M Karvanen, K Pontikis, B Jansson, et al. Application of a loading dose of colistin methanesulfonate in critically ill patients: population pharmacokinetics, protein binding, and prediction of bacterial kill. AAC. 2012;56:4241-9.
- Li J, Milne RW, Nation RL, Turnidge JD and Coulthard K. Stability of colistin and colistin methanesulfonate in aqueous media and plasma studied by high-performance liquid chromatography. AAC. 2003;47:1364-70.
- Li J, Milne RW, Nation RL, Turnidge JD, Smeaton TC and Coulthard K. Pharmacokinetics of colistin methanesulphonate and colistin in rats following an intravenous dose of colistin methanesulphonate. JAC. 2004;53:837-40.
- Warunee S. Polymixin re-visited: carbapenem resistant Gram-negative bacteria. Texas: The University of Texas. 2010;2-16.
- Kasiakou SK, Michalopoulos A and Soteriades ES. Combination therapy with intravenous colistin for management of infections due to multidrug-resistant Gram-negative bacteria in patients without cystic fibrosis. AAC. 2005;49:3136-46.
- Spellberg B, Blaser M and Guidos RJ. Combating antimicrobial resist- ance: policy recommendations to save lives. CID. 2011;52(Suppl 5):S397-428.
- Beceiro A, Moreno A and Fernandez N. Biological cost of different mech- anisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. AAC. 2014;58:518-26.
- Couet W, Grégoire N and Gobin P. Pharmacokinetics of colistin and colistimethate sodium after a single 80-mg intravenous dose of CMS in young healthy volunteers. CPT. 2011;89:875-9.
- Nation RL and Li J. Colistin in the 21st century. COID. 2009;22(6):535-43.
- Velkov T, Roberts K, Nation R, Ompson P and Li J. Pharmacology of polymyxins: new insights into an “old” class of antibiotics. FM. 2013;8(6):711-24.
- Yahav D, Farbman L, Leibovici L and Paul M. Colistin: new lessons on an old antibiotic. CMI. 2012;18:18-29.
- Evans ME, Feola DJ and Rapp RP. Polymyxin B sulfate and colistin: Old antibiotics for emerging multiresistant gram-negative bacteria. APT. 1999;33:960-7.
- Powers JP and Hancock RE. e relationship between peptide structure and antibacterial activity. Peptides. 2003;24(11):1681-91.
- Lexicomp Online, Lexi-Drug®, Hudson, Ohio: Lexi-Comp, Inc. 2016.
- Nation RL, Velkov T and Li J. Colistin and polymyxin B: Peas in a pod, or chalk and cheese? CID. 2014;59:88-94.
- Falagas ME, Rafailidis PI and Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options. DRU. 2010;13:132-854.
- Sampson TR, Liu X and Schroeder MR. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. AAC. 2012;56:5642-9.
- Karvanen M. Optimization of Colistin Dosage in the Treatment of Multiresistant Gram-negative Infections. AUU. 56 pp.
- Kwa AL, Tam VH and Falagas ME. Polymixins: a review of the current status including recent developments. AAMS. 2008;37:870-3.
- Fern´andez C. ´ Alvarez-Ortega and I Wiegand. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. AAC. 2013;57(1):110-9.
- SA Loutet, LE Mussen, RS Flannagan and MA Valvano. A two-tier model of polymyxin B resistance in Burkholderia cenocepacia. EMR. 2011;3(2):278-85.
- Velkov P, E Thompson, RL Nation and J Li. Structureactivity relationships of polymyxin antibiotics. JMC. 2010;53(5):1898-916.
- Nikaido. Molecular basis of bacterial outer membrane permeability revisited. MMBR. 2003; 67(4):593-656.
- Cannatelli A, D’Andrea MM and Giani T. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. AAC. 2013;57:5521-6.
- Napier BA, Burd EM, Satola SW. Clinical use of colistin induces cross resistance to host antimicrobials in Acinetobacter baumanii. MBIO. 2013;4:1-4.
- Skov RL and Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. ES. 2016;21.
- Poirel L, Jayol A and Bontron S. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. TJAC. 2015;70:75-80.
- Liu Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet. 2016;16:161.
- Skov RL and Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. ES. 2016;21.
- Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Tu¨ rkoglu S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. JAC. 2015;70:75-80.
- Olaitan AO, Morand S and Rolain J-M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. FM. 2014;5:643.
- Falagas ME, Rafaildis PI and Matthaiou DK. Resistance to polymixins: mechanisms, frequency and treatment options. DRU. 2010;13:132-8.
- Trent MS, Ribeiro AA, Lin S, Cotter RJ and Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-Larabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. JBC. 2001;276:43122-31.
- Needham BD and Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. NRM. 2013;11:467-81.
- Petrou VI, Herrera CM, Schultz KM, Clarke OB, Vendome J, Tomasek D, et al. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science. 2016; 351:608-12.
- Fage CD, Brown DB, Boll JM, Keatinge-Clay AT and Trent MS. Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni. ACD. 2014;D70:2730-9.
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. MMBR. 2003; 67:593-656.
- Stojanoski V, Sankaran B, Prasad V, Poirel L, Nordmann P and Palzkill T. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC B. 2016;14(1):81.
- Wanty C, Anandan A, Piek S, Walshe J, Ganguly J, Carlson RW, et al. The structure of the Neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymixin. JMB. 2013;425:3389-402.
- Hinchliffe P, Yang Q, Portal E, Young T, Li H, Catherine L. Tooke, et al. Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1. SR. 2017;7:39392.
- Schwarz S and Johnson AP. Transferable resistance to colistin: a new but old threat. JAC. 2016;71: 2066-70.
- Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of Plasmid Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. LID. 2016;16(2):161-8.
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P and Goossens H. Colistin Resistance Gene mcr-1 Harboured on a Multidrug Resistant Plasmid. LID. 2016;16(3):283-4.
- Xavier BB, Lammens C, Butaye P, Goossens H and Malhotra-Kumar S. Complete Sequence of an IncFII Plasmid Harbouring the Colistin Resistance Gene mcr-1 Isolated From Belgian Pig Farms. J Antimicrob Chemother. 2016;71(8):2342-4.
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. ES. 2016; 21(27):30280.
- Berrisford JM, Baradaran R and Sazanov LA. Structure of Bacterial Respiratory Vomplex I. BBA. 2016;1857(7):892-901.
- World Health Organization (WHO). 2017.
- Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. IPID. 2012;976273.
- Nang SC, Li J and Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. CRM. 2019;45(2):131-61.
- Shah AA, Hasan F, Ahmed S and Hameed A. Extended-spectrum beta-lactamases (ESbLs): characterization, epidemiology and detection. CRM. 2004;30:25-32.
- El Salabi A, Walsh TR and Chouchani C. Extended spectrum β-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. CRM. 2013; 39(2):113‐122.
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. CMR. 2001;14:933-51
- Livermore DM, Warner M, Mushtaq S, Doumith M, Zhang J and Woodford N. What remains against carbapenem resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. IJAA. 2011;37:415-9.
- Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. Agents of last resort: polymyxin resistance. IDCNA. 2016;30:391-414.
- Ye H, Li Y, Li Z, Gao R, Zhang H, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio. 2016;7:e00177.
- Chen K, Chan EW, Xie M, Ye L, Dong N and Chen S. Widespread distribution of mcr-1-bearing bacteria in the ecosystem, 2015 to 2016. Euro surveillance: bulletin Europeen sur les maladies transmissibles. ECDB. 2017;22(39).
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. ES. 2016; 21(27):30280.
- McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, et al. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. AAC. 2016; 60:4420-1.
- Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RN and Flamm RK. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program during 2014-2015. AAC. 2016;60(9):5623-4.
- Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, et al. Detection of mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae (CRE) from different hospitals in China. AAC. 2016;60(8):5033-5.
- Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, et al. Colistin resistance gene mcr-1 in extendedspectrum -lactamase-producing and carbapenemase-producing Gramnegative bacteria in Germany. LID. 2016;16:282-3.
- Poirel L, Kieffer N, Liassine N, Thanh D and Nordmann P. Plasmidmediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. LID. 2016;16(3):281.
- Zhou YF, Tao MT, Feng Y, Yang RS, Liao XP, Liu YH, et al. Increased activity of colistin in combination with amikacin against Escherichia coli co-producing NDM-5 and MCR-1. JAC. 2017;72:1723-30.
- Oz T, Guvenek A, Yildiz S, Karaboga E, Tamer YT, Mumcuyan N, et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. MBE. 2014;31:2387-401.
- Lazar V, Pal Singh G, Spohn R, Nagy I, Horvath B, Hrtyan M, et al. Bacterial evolution of antibiotic hypersensitivity. MSB. 2013;9:700.
- Geller BL, Deere JD, Stein DA, Kroeker AD, Moulton HM and Iversen PL. Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. AAC. 2003;47:3233-9.
- Deere J, Iversen P and Geller BL. Antisense phosphorodiamidate morpholino oligomer length and target position effects on gene-specific inhibition in Escherichia coli. AAC. 2005;49:249-55.
-

