D.SAP, a Formulation Derived from Apples, Showed the Potential to Reduce Inflammation in COVID-19 Infected Patients
Hossein Dezhakam1,2, Ani Dezhakam1,2, Amin Dezhakam1,2, Shani Dezhakam1,2 and Arvin Haghighatfard 1,*
1Khwarizmi Institute of science and technology, Qeshm island, Hormozgan, Iran
2Congress 60 non-governmental organization, Iran
Received Date: 19/09/2024; Published Date: 04/11/2024
*Corresponding author: Arvin Haghighatfard, Khwarizmi Institute of science and technology, Qeshm island, Hormozgan, Iran
Abstract
Introduction: The SARS-CoV-2 virus is a respiratory virus that causes severe acute respiratory disorder with an unknown mortality rate. It has been reported that excessive inflammation in COVID-19-infected patients could lead to unfavorable immune reactions such as cytokine storms, which could even cause death. The search for therapeutic strategies to suppress or modulate inflammation in COVID-19 is an important goal not only for COVID-19 infection but for other acute respiratory infections. We aimed to evaluate the effects of using an apple-based formulation called Dezhakam sap (D.SAP) in the reduction of COVID-19-associated inflammation response of individuals with COVID-19 infection.
Methods: We recruited 210 patients with COVID-19 infection who had used D.SAP for at least 12 months before the viral infection and 350 patients with COVID-19 infection who had not used D.SAP. A matched control healthy group with no history of COVID-19 infection, including 1080 subjects used for comparison with infected groups. Blood samples obtained from all subjects' RNA were extracted from blood and cDNA was synthesized from RNAs. Then all subjects were examined for inflammation markers in blood samples using Real-time PCR.
Results: Results showed significant alterations in mRNA level of inflammation factor genes in patients with D.SAP usage reputation compared to COVID-19-infected subjects without using D.SAP.
Conclusion: Results showed the effectiveness of specially formulated apple vinegar D.SAP as a natural anti-inflammatory agent. It seems that the anti-inflammation effects of D.SAP users may related to the antioxidant effects of polyphenolic compounds of apples that were maintained during the process of D.SAP production. Reduction of inflammation markers may increase the survival rate in patients with severe reactions of the immune system in COVID-19 infection.
Keywords: COVID-19; Apple; D.SAP; Inflammation; Gene expression
Introduction
The SARS-CoV-2 virus is a positive-sense RNA virus from the Coronavirus family that causes a severe acute lethal respiratory disorder called COVID-19. While the lethal rate of COVID-19 is still unclear, patients who required intensive care show higher levels of excessive systemic inflammation, pro-inflammatory factor release, hemostasis, and coagulation system activation along with severe cytotoxic effects and cytokine cascade [1]. Systemic inflammatory activation caused by COVID-19 infection could lead to the disruption of several molecular and cellular homeostatic mechanisms, favoring toxicity and even organ failure [2].
Several biomarkers have been such as C-reactive protein, lactate dehydrogenase, and D-dimer evaluated to predict the excessive inflammatory response in COVID-19 infection [3,4]. Neutrophia, lymphopenia, and thrombocytopenia, along with associated hematological indexes, have been reported to predict excessive inflammation and severe disease and mortality in patients infected with COVID-19 [5]. Calculation of the systemic inflammatory index (SII) [6], in patients with COVID-19 showed a significant correlation between SII and severe disease and mortality rate [7].
It has been reported that COVID-19 infection is strongly associated with immune dysregulation and cytokine storm. Biomarker analysis to achieve predictive markers of immune response severity in COVID-19 patients revealed decreased CD3+, CD4+, and CD8+ T cells and increased neutrophils in circulation, exhibiting upregulated neutrophil-to-lymphocyte and neutrophil-to-CD8+ T cell ratio [8]. In addition, dramatically increase of IL-6, TNF-α, IL-1β, IL-18, IL-12/IL-23p40, IL-10, Tim-3, IL-8, neutrophil extracellular trap–related proteinase 3, and S100A8/A9 were suggested as a potential predictive marker for COVID-19 severity and COVID-19 progression along with potential targets for novel therapeutic intervention for patients [8,9]. Also, cytokine storm as an important cause of severe inflammatory response and mortality of COVID-19 patients is associated with level elevation of pro-inflammatory cytokines, such as IL-6, IL-8, IL-1β, G-SCF, GM-SCF, IP10, MCP-1, MCP-3, IL-1ra, MIP1α, and TNF-α [10-12].
Previous studies have documented the antimicrobial effects of polyphenolic compounds in apple cider vinegar. On the other hand, the antioxidant effects of polyphenolic compounds and probiotic bacteria such as lactobacillus serotypes in apple cider vinegar have anti-inflammatory effects that may influence the immune system functions. Dezhakam sap (D.SAP) is an apple-based formulation that is produced by a traditional method in Iran. Because of the selection process of apples and the special traditional production of D.SAP, antioxidants, probiotic compounds (especially lactobacillus strains), micronutrients and cellulose fiber of the D.SAP are dramatically higher than apple vinegar. It has been reported that the contents of D.SAP may have a potentially positive effect on the treatment of acute respiratory disorders such as influenza and the SARS-CoV-2 virus regarding its anti-inflammation effects [13].
The present study aimed to evaluate the inflammatory-preventing effect of long-term oral usage of D.SAP (an especially formulated apple-based production) on patients with COVID-19 infection. The expression level analysis of a short list of inflammation system marker genes extracted from the previous studies in peripheral blood was performed to examine D.SAP anti-inflammatory effects.
Material, Methods, and Methodology
Subject recruitment and grouping:
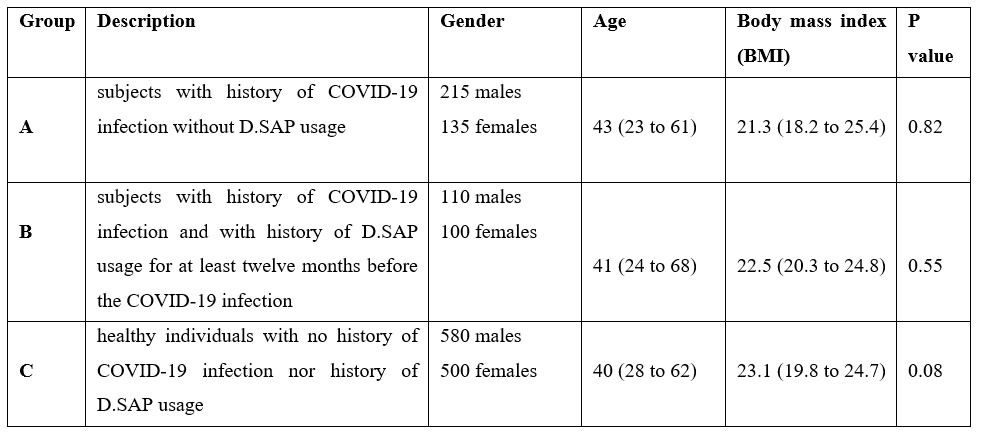
Subjects of the present study included two groups of previously COVID-19-infected individuals. The A group included subjects with a history of COVID-19 infection without D.SAP usage, the B group included subjects with a history of COVID-19 infection and with a history of D.SAP usage for at least twelve months before the COVID-19 infection, and group C included healthy individuals with no history ofCOVID-19 infection nor history of D.SAP usage. Demographic data of all groups were presented in Table 1. Informed consent was obtained from all participants before the sampling.
Blood sampling and RNA extraction:
The collection of blood samples took place between 10.00 and 12:00 AM using PAXgene vacutainer blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland). RNA was extracted from blood samples immediately after samplings according to standard protocols using RNA Purification kit (GeneJET™ RNA Purification Kit#K0732, Thermo Scientific - Fermentas, Latvia). The manufacturer's protocol recommends removing any genomic DNA contamination from extracted RNA that has been treated with DNase Treatment and Removal Reagents (DNase I, RNase-free (#EN0521) Fermentas, Latvia). After removing of genomic DNA, the quality and quantity of extracted RNA were examined by Agarose horizontal gel electrophoresis and UV- spectroscopy respectively.
cDNA synthesis:
Synthesis of cDNA was performed by Transcription First Strand cDNA Synthesis Kit (RevertAid Premium First Strand cDNA Synthesis Kit #K1652, Thermo Scientific -Fermentas, Latvia) based on the manufacturer’s guidelines.
Expression analysis of inflammation genes:
Specific primers were created using the oligo7 software and then promoted on the NCBI website. Standard curves for each gene were prepared using serial dilutions (1: 4) of pooled cDNA from total extracted RNA. In each experiment, the R2 value of the standard curve was more than 0.99, and no-template control assays showed no detectable signal. The Quantitative Real-time PCR with a triplicate strategy was performed and the efficacy of the PCR reaction was calculated using Lin-Reg PCR software (Amsterdam, Netherlands). Real-time PCR was conducted using Maxima SYBR Green/ROX qPCR Master Mix, #K0221 (Thermo scientific -Fermentas, Latvia) and CFX96 Touch Real-Time PCR Detection System (BIO-RAD, California, United States).
Statistical analysis:
A P value less than 0.05 was chosen as the statistical significance level, and the descriptive data is represented as mean <unk> SD (range). Normal distribution evaluation for continuous variables was conducted with the Kolmogorov-Smirnov test. The one-way ANOVA and independent Student's t-test were utilized to compare multiple groups. Determination of the correlations between variables examined by Pearson correlation analysis. The Bonferroni correction test was used to conduct multiple comparison corrections. Version 24 of Statistical Package for the Social Sciences (SPSS) software used for statistical analysis.
Table1: Group's names and demographic data.
Results
Demographic data including age, gender and body mass index (BMI) have been presented in Table 1. Statistical analysis showed no significant difference in demographic data between groups.
Gene expression results:
Significant downregulation of IL-6, IL-1β, TNF-α, IL-18, IL-12, IL-10, IL-8, and HAVCR2 genes in group B compared to group A were detected. Also, significant upregulation of CD3, CD4, and CD8 genes in group B compared to group A was determined. Detailed gene expression ratio and statistical data for each gene have been presented in Tables 2 and 3 respectively. Correlation analysis showed significant direct correlation between down expression of IL-6 and IL-1β (R= 0.66, p-value: 0.002), direct correlation of IL-6 and TNF-α (R= 0.63, p-value: 0.002), direct correlation of IL-18 and IL-18 (R= 0.58, p-value: 0.004), and indirect correlation of IL-6 and CD4 (R= - 0.55, p-value: 0.004).
Table 2: Gene expression level of target genes in peripheral blood sample.
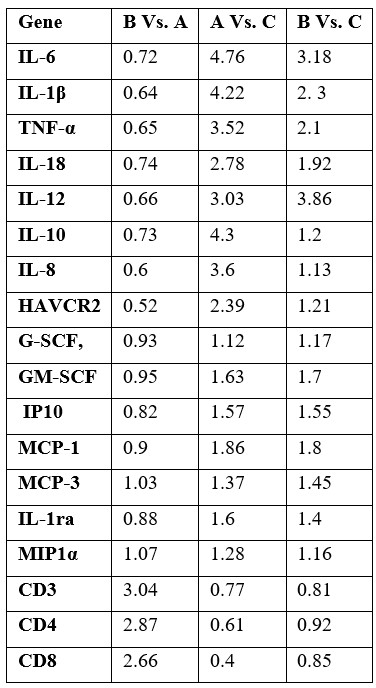
Group A: subjects with history of COVID-19 infection without D.SAP usage, group B: subjects with history of COVID-19 infection and with history of D.SAP usage for at least twelve months before the COVID-19 infection, group C: healthy individuals with no history of COVID-19 infection nor history of D.SAP usage.
Table 3: Statistical analysis of target genes' mRNA level in blood samples in each group.
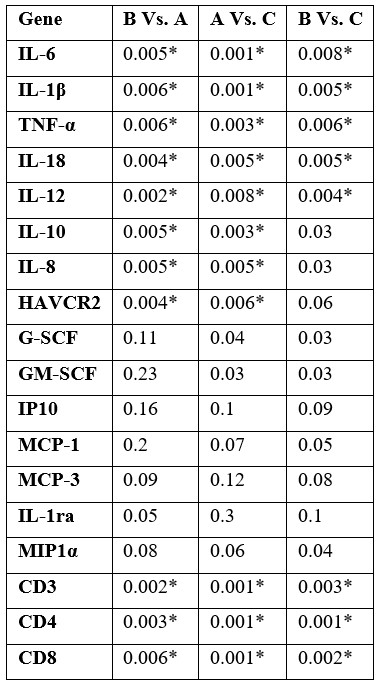
Group A: subjects with history of COVID-19 infection without D.SAP usage, group B: subjects with history of COVID-19 infection and with history of D.SAP usage for at least twelve months before the COVID-19 infection, group C: healthy individuals with no history of COVID-19 infection nor history of D.SAP usage. The * sign used for significant corrected p-values.
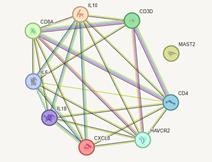
Figure 1: A graphical description of protein-protein interaction between significantly differentially expressed genes in Group B Vs. Group A drew with String data base (https://string-db.org/cgi/).
Discussion
IL6, IL1b, IL-18, IL-12, IL-10, IL-8, HAVCR2, and TNF-α, which showed over-expression in the D.SAP users group, are major inflammatory markers and proinflammatory cytokines that may be used in the determination of inflammation response of patients with acute respiratory diseases and associated concomitant disorders such as arterial hypertension and metabolic disorders, etc [14]. SARS-CoV-2 may cause a cytokine storm that results in the chemotaxis of neutrophils through IL-8/IL-6, leading to severe inflammatory responses [15]. Coronavirus infection associated with pyroptosis is related to an increase of IL-1β and IL-18 mostly produced by macrophages in the serum of COVID-19 patients, especially in severe and hospitalized cases [16]. The discovery of severe inflammasome activation during SARS-CoV-2 infection has resulted in a cytokine storm and pyroptosis in hematopoietic stem cells [17]. IL-18 upregulation in turn could cause an increase in ferritin concentration which is strongly correlated with the severity of infection and increase the rate of heart failure in COVID-19 patients [18]. It has been reported that overexpression and dysfunction of HAVCR2 of T cells are associated with the onset of several autoimmune diseases and exacerbation of clinical symptoms in viral infections [19,20]. The B group's significant decrease in HAVCR2 may suggest a decreased inflammation response in COVID-19 patients with a history of D.SAP usage.
On the other hand, several lines of evidence indicated that significant reduction in expression of CD3, CD4, and CD8 genes in COVID-19 patients with moderate infection and under-medication groups [21,22]. Previous studies, also reported that low count of CD3+, CD4+, and CD8+ T cells in severe cases of COVID-19 infection and regulation of these T cells in recovered COVID-19 patients compared to the healthy subjects [23-25]. In addition, CD4+ and CD8+ T cell responses have the main role in the resolution of SARS-CoV-2 infection such as modulating disease severity, viral load reduction, and COVID-19 vaccine immunity production [26-28]. Interestingly, the downregulated genes (IL6, IL1b, IL-18, IL-12, IL-10, IL-8, HAVCR2, and TNF-α) and downregulated genes (CD3, CD4, and CD8) in D.SAP users have a strong relation in protein level and could function in shared gene ontologies and pathways. Figure 1 illustrates the correlation between genes that are differentially expressed in D.SAP users of group B.
Previous reports about D.SAP usage of the COVID-19 rodent model showed that pretreatment with D.SAP could cause low pathogenicity and viral load, as well as easier treatment and recovery in rodents [13]. It has been suggested that probiotics and anti-oxidant compounds could lead to anti-inflammation activities that in turn may help the immune system to better respond to influenza-like acute respiratory infections [29]. Investigation of gut bacteria effects on the severity of COVID-19 infection showed the upregulation of IL-18, in COVID-19 patients, could lead to gut microbiota dysbiosis due to severe infection that in turn, could cause intestinal infection in COVID-19 patients [30]. Clinical studies in autoimmune disease revealed the administration of probiotics especially some of Lactobacillus strains could cause the reduction and/or inhibition of inflammatory mediators such as TNF α, IL‐6, and IL‐1β [31]. Strengthening the immune system using prophylactic-probiotics approaches to reduce the severity of COVID-19 infection has several benefits including balancing the composition of human gut microflora, strengthening gut barrier function, and preventing inflammatory response [32].
While the mechanisms of SARS-CoV-2-induced inflammatory response are not completely clarified, it seems that using probiotic and antioxidant-enriched compounds such as apple-based production like D.SAP in the long term could reduce the possibility of cytokine storm and other major and life-threatening inflammatory reactions in COVID-19 patients.
Ethics Statement: This research was approved by the Central Ethical Committee of the Islamic Azad University with the approval number 39662.
Conflict of interest: The authors declare that they have no conflict of interest.
References:
- Andrade SA, de Souza DA, Torres AL, de Lima CF, Ebram MC, Celano RM, et al. Pathophysiology of COVID-19: critical role of hemostasis. Frontiers in cellular and infection microbiology, 2022; 12: 896972.
- Silva MJ, Ribeiro LR, Gouveia MI, Marcelino BD, Santos CS, Lima KV, et al. Hyperinflammatory response in COVID-19: a systematic review. Viruses, 2023; 15(2): 553.
- Zinellu A, Paliogiannis P, Carru C, Mangoni AA. Serum hydroxybutyrate dehydrogenase and COVID-19 severity and mortality: a systematic review and meta-analysis with meta-regression. Clinical and Experimental Medicine, 2022: 1-10.
- Qin R, He L, Yang Z, Jia N, Chen R, Xie J, et al. Identification of parameters representative of immune dysfunction in patients with severe and fatal COVID-19 infection: a systematic review and meta-analysis. Clinical Reviews in Allergy & Immunology, 2023; 64(1): 33-65.
- Wei T, Li J, Cheng Z, Jiang L, Zhang J, Wang H, et al. Hematological characteristics of COVID‐19 patients with fever infected by the Omicron variant in Shanghai: A retrospective cohort study in China. Journal of Clinical Laboratory Analysis, 2023; 37(1): e24808.
- Hu B, Yang XR, Xu Y, Sun YF, Sun C, Guo W, et al. Systemic immune-inflammation index predicts prognosis of patients after curative resection for hepatocellular carcinoma. Clinical Cancer Research, 2014; 20(23): 6212-6222.
- Zinellu A, Scano V, Masotto E, De Riu G, Vaira LA, Carru C, et al. The systemic inflammation index on admission is independently associated with length of stay in hospitalized COVID-19 patients. Minerva Respir Med, 2021; 60(3): 65-72.
- Huang W, Li M, Luo G, Wu X, Su B, Zhao L, et al. The inflammatory factors associated with disease severity to predict COVID-19 progression. The Journal of Immunology, 2021; 206(7): 1597-1608.
- Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature medicine, 2020; 26(6): 842-844.
- Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerging microbes & infections, 2020; 9(1): 761-770.
- Yang Y, Shen C, Li J, Yuan J, Wei J, Huang F, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. Journal of Allergy and Clinical Immunology, 2020; 146(1): 119-127.
- Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clinical infectious diseases, 2020; 71(15): 762-768.
- Dezhakam H, Dezhakam A, Haghighatfard A. D. SAP, an apple-based formulation could treat SARS-CoV-2 infection, and reduce associated inflammatory responses in COVID-19 infected mice model, Archives of Infect Diseases & Therapy, 2022; 6(2): 183-188.
- Chiok K, Hutchison K, Miller LG, Bose S, Miura TA. Proinflammatory Responses in SARS-CoV-2 and Soluble Spike Glycoprotein S1 Subunit Activated Human Macrophages. Viruses, 2023; 15(3): 754.
- Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. The Journal of clinical investigation, 2020; 130(5): 2202-2205.
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Molecular cell, 2002; 10(2): 417-426.
- Ratajczak MZ, Kucia M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells. Leukemia, 2020; 34(7): 1726-1729.
- Soundravally R, Agieshkumar B, Daisy M, Sherin J, Cleetus CC. Ferritin levels predict severe dengue. Infection, 2015; 43: 13-19.
- Ju Y, Hou N, Meng J, Wang X, Zhang X, Zhao D, et al. T cell immunoglobulin-and mucin-domain-containing molecule-3 (Tim-3) mediates natural killer cell suppression in chronic hepatitis B. Journal of hepatology, 2010; 52(3): 322-329.
- Yang X, Jiang X, Chen G, Xiao Y, Geng S, Kang C, et al. T cell Ig mucin-3 promotes homeostasis of sepsis by negatively regulating the TLR response. The Journal of Immunology, 2013; 190(5): 2068-2079.
- Aljabr W, Al-Amari A, Abbas B, Karkashan A, Alamri S, Alnamnakani M, et al. Evaluation of the levels of peripheral CD3+, CD4+, and CD8+ T cells and IgG and IgM antibodies in COVID-19 patients at different stages of infection. Microbiology spectrum, 2022; 10(1): e00845-21.
- Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Reports Medicine, 2021; 2(7).
- Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clinical infectious diseases, 2020; 71(15): 762-768.
- Chen G, Wu DI, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. The Journal of clinical investigation, 2020; 130(5): 2620-2629.
- Calvet J, Gratacós J, Amengual MJ, Llop M, Navarro M, Moreno A, et al. CD4 and CD8 lymphocyte counts as surrogate early markers for progression in SARS-CoV-2 pneumonia: a prospective study. Viruses, 2020; 12(11): 1277.
- Moderbacher CR, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell, 2020; 183(4): 996-1012.
- Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell reports, 2021; 34(6).
- Breton G, Mendoza P, Hägglöf T, Oliveira TY, Schaefer-Babajew D, Gaebler C, et al. Persistent cellular immunity to SARS-CoV-2 infection. Journal of Experimental Medicine, 2021; 218(4): e20202515.
- Ayyanna R, Ankaiah D, Arul V. Anti-inflammatory and antioxidant properties of probiotic bacterium Lactobacillus mucosae AN1 and Lactobacillus fermentum SNR1 in Wistar albino rats. Frontiers in microbiology, 2018; 9: 425828.
- Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Medicine in Microecology, 2020; 5: 100023.
- Akour A. Probiotics and COVID‐19: is there any link? Letters in applied microbiology, 2020; 71(3): 229-234.
- Singh K, Rao A. Probiotics: A potential immunomodulator in COVID-19 infection management. Nutrition Research, 2021; 87: 1-2.

