Strength of Association Between Generalized/Nonspecific Covid-19 Signs & Symptoms With SARS-COV 2 Specific ORF, N, E Genes Identified Through Real Time PCR
Muhammad Arif*, Muhammad Abdullah, Sheikh Ahmed, Mir Abdul Qadir, Ehsan Ahmed Larik, Ujala Naseer Baloch, Zakir Hussain and Mirza Zeeshan Iqbal Baig
Department of Health, FELTP Pakistan, Quetta, Pakistan
Received Date: 25/05/2023; Published Date: 25/09/2023
*Corresponding author: Mohammad Arif, Department of Health, FELTP Pakistan, Quetta, Pakistan
Abstract
Background: Constant mutation in the SARS-COV2 virus genetic material is resulting in the appearance of new variants frequently hence the overall virulence, treatment resistance, replication modalities, transmissions rates and COVID-19 signs & symptoms are all changing regularly.
Methodology: From 1 January 2021 to 30 August 2022, the clinical lab at Fatima Jinnah General & Chest Hospital Quetta, Balochistan, determined a total of 3375 individuals to be COVID-19 positive because RT-PCR detected ORF, N, and E genes or their various Bi & Tri combinations in their samples. A questionnaire-based interview was conducted with each participant during sample collection. Body temperature more than 370c was recorded as Fever/Chill. Age, Comorbidities, A-symptomatic individuals & Vaccination status were all neglected during this study. Frequency tables were generated using MS-excel 2016, while Odds ratios were calculated using Chi-square test of association whereby 2x2 contingency tables between Mono, Bi & Tri combinations for ORF, N & E genes were cross associated with various generalized nonspecific COVID -19 signs and symptoms using Epi-info software. Absence of Genetic sequencing was the major limitation.
Results: The study showed that individually the presence of ORF gene was found to be strongly associated “ Shortness of Breath/Difficulty in Breathing”, Diarrhea, Head ache & Vomitting. While the presence of N-gene was found to be strongly associated with Loss of smell & taste, Head ache,Presistant Chest Pain & Bluish lips/Face. Where as the presence of E-gene was found to be strongly associated with Cough, Shortness of breath/ Difficulty in breathing, Sore throat, Diarrhea, Head ache & Laziness. In addition, the study also found that different Bi & Tri combinations of ORF, N & E genes in a COVID-19 positive patient expressed generalized non-specific COVID-19 signs & symptoms differently.
Discussion & Conclusion: The presence of various SARS-COV2 genetic markers significantly alters the clinical presentation of COVID-19.
Keywords: RT-PCR; SARS-COV2; ORF N & E Genes; Association
Introduction
There were 41,409 people in all documented, from at least 23 different nations, with 26 different clinical presentations. Six symptoms—fever (58.66%), cough (54.52%), dyspnea (30.82%), malaise (29.75%), weariness (28.16%), and sputum/secretion (25.33%)—had a general prevalence more than or equal to 25%. Other prevalent symptoms included headache (12.17%), chest discomfort (11.49%), diarrhea (9.59%), sneezing (14.71%), sore throat (14.41%), rhinitis (14.29%), goosebumps (13.49%), dermatological signs (20.45%), anorexia (20.26%), myalgia (16.9%), and rhinitis. The manifestations of dermatology were only documented in one study. Hemoptysis was the least common indication or symptom (1.65%). The three most common symptoms in trials involving more than 100 patients were dyspnea (30.64%), cough (54.21%), and fever (57.93%) [1-13].
Certain symptoms, such as dyspnea, fever, cough, and headache, are generally nonspecific for SARS-CoV2. Patients who are asymptomatic may have mild forms of infection, while others may have severe pneumonia that can be fatal [2-3].
The initial symptoms of the illness were the trio of fever, coughing, and shortness of breath. Later, the US Center for Disease Control and Prevention (CDC) expanded this list to include chills, headache, sore throat, muscle discomfort, and loss of taste or smell (neurological manifestations) [4-15].
Any possible association between generalized nonspecific COVID-19 signs and symptoms with SARS-COV 2 specific ORF, N & E genes would open up a new door to research in future.
Literature Review
It is certain that age manifest the clinical sings & symptoms of COVID-19 differently. According to one study, people over 60 have more bilateral lobe lesions, greater levels of inflammatory markers, and higher blood urea nitrogen levels. Patients who are older than 60 have a higher risk of respiratory failure and prolonged illness courses. The intensity is, however, less severe in people under 60 [5-14].
According to one additional study that claims a total of 72,314 verified cases in China the majority of the patients (87%) are between the ages of 30 and 79. There were no fatalities among those under the age of nine. However, the case-fatality rate (CFR) is 8.0% for those aged 70 to 79 and 14.8% for people 80 years of age and older. The CFR is 10.5, 7.3, 6.3, 6.0, and 5.6%, respectively, for patients with various concomitant diseases, including cancer, chronic respiratory disease, diabetes, cardiovascular disease, and hypertension. According to these findings, COVID-19 patients with comorbid conditions have higher fatality rates than those who don't have underlying diseases [6-15].
Coronaviruses have genomes that range in size from 26 to 32 kilobases and have a variety of open reading frames (ORFs) [7]. Coronaviruses have a varying number of open reading frames in their genomes, which range in size from 26 to 32 kilobases (ORFs). The spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N) are the four major structural proteins and the eight accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14) are situated in the 3′-terminus of the SARS-CoV-2 genome [8-16,17].
The SARS-CoV-2 was found to be more related to two SARS-like bat CoVs from Zhoushan in eastern China, bat-SL-CoVZC45 and bat-SL-CoVZXC21, than to the SARS-CoV and the MERS-CoV, according to analysis of the genome from the samples of nine patients. Laboratory specific detection Next-generation sequencing or real-time reverse transcriptase-polymerase chain reaction (RT-PCR) techniques for the SARS-CoV-2 virus were developed as a result of the isolation of the causative agent and determination of its partial genomic sequence [9-20].
The Open Reading Frame (ORF) segments in the SARS COV 2 virus genome encode structural and non-structural proteins [8]. The ORF1a/b gene in SARS COV 2 nucleic acid is used for diagnostics by RT PCR. It produces non-structural proteins (nsp1–16), which are necessary for the viral genome's maintenance and replication apparatus. Adaptive mutations in ORF1a/b may boost viral replication or increase treatment resistance, hence increasing virulence [10]. The “N” genome encodes the “N” structural protein which participates in a number of viral genome-related functions, such as viral genome signaling, viral replication, and host cell immunity to viral infections [11]. Similarly, the “E” genome encodes for the “E” structural protein Through interactions with the host cell membrane protein, the E protein participates in the viral growth and maturation stages [12-25].
From the very first case the SARS-COV 2 infection, a continuous mutation is reported in the virus as a result of which new variants popup frequently hence the overall virulence, treatment resistance, replication modalities & transmissions rates are all changing regularly hence in these circumstances it is of great importance to frequently monitor the symptomology of COVID-19. Currently no published literature has assessed the relationship of COVID-19 symptomology (i.e., COUGH, SHORTNESSS OF BREATH/DIFFICULTY IN BREATHING, Fever/Chills, NEW MUSCLE/BODY ACHE, SORE THROAT, LOSS OF SMELL & TASTE, DIARRHEA, HEAD ACHE, NAUSEA, VOMITTING, RUNNY NOSE, PERSISTANT CHEST PAIN, LAZINESS, BLUISH LIPS/FACE) with the ORF, N & E genes which are few of the diagnostic markers assessed for SARS COV-2 infection assessed during real-time PCR test.
Methodology
From 1 January 2021 to 30 August 2022, the clinical lab at Fatima Jinnah General & Chest Hospital Quetta, Balochistan, determined a total of 3375 individuals to be COVID-19 positive because RT-PCR detected ORF, N, and E genes or their various Bi & Tri combinations in their samples. A questionnaire-based interview was conducted with each participant during sample collection. Body temperature more than 370c was recorded as Fever/Chill. Age, Comorbidities, A-symptomatic individuals & Vaccination status were all neglected during this study. Frequency tables were generated using MS-excel 2016, while Odds ratios were calculated using Chi-square test of association whereby 2x2 contingency tables between Mono, Bi & Tri combinations for ORF, N & E genes were cross associated with various generalized nonspecific COVID -19 signs and symptoms using Epi-info software. Absence of Genetic sequencing was the major limitation.
Results
From 01 January 2021 till 30th August 2022 a total of 3375 local residents across Balochistan were declared to be COVID-19 positive by the RT-PCR. 48% (n=1620) individuals out of 3375 study participants were found to be positive for all the three major diagnostic markers of SARS-COV 2 (i.e. ORF + N + E). Similarly, the RT-PCR reports of 12% (n=405) study participants were positive for only N & E (N + E) genes together. Moreover 13% (n= 438) study participants were positive only for ORF & N (ORF + N) genes together. While 10% (n=337) study participants were positive only for ORF & E (ORF+E) genes combination. It was also seen that 08% (n=270) study participants were positive only for ORF gene, 06% (n=203) study participants expressed only “N” gene while only 03% (n=102) of the study participants were positive only for “E” gene only as shown below:
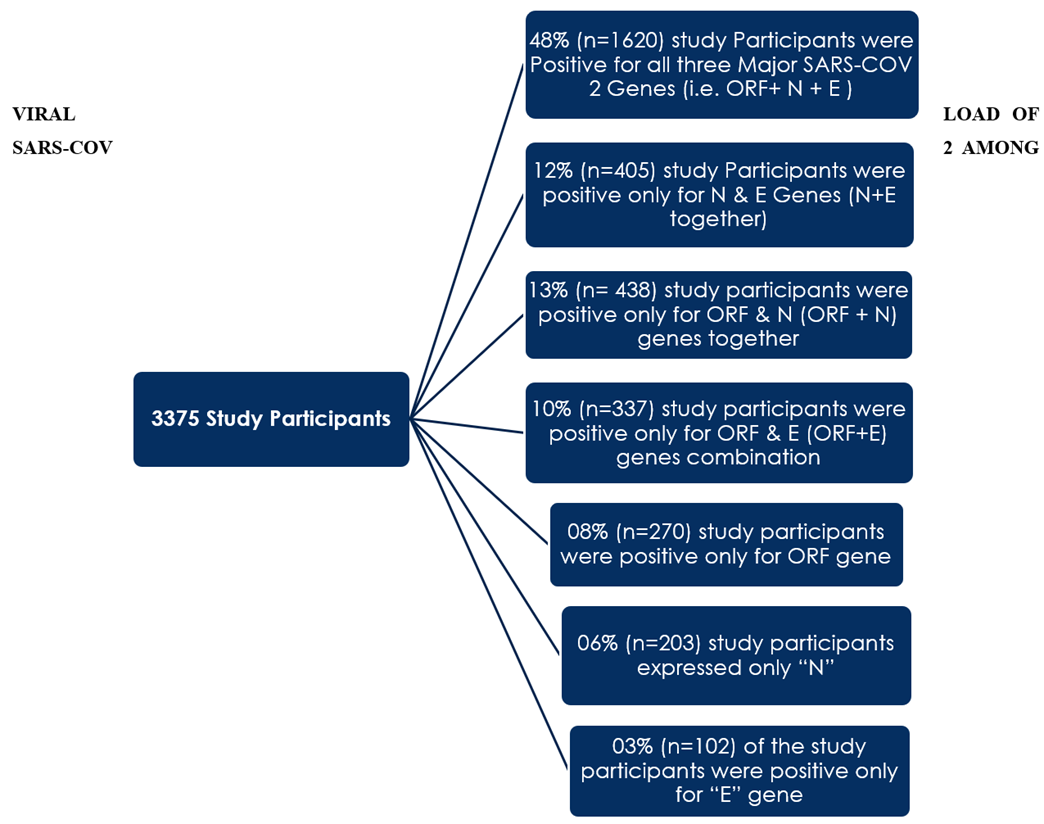
Figure 1: Sub classification of the study participants based on results of RT-PCR reports.
Viral Load of SARS-COV2 Among Study Participants:
If a sample gets positive and show presence of ORF, N & E genes in any combination on lesser RT-PCR cycles i.e., below 21 cycles is generally believed to possess high viral load while if a sample becomes positive at 21 or beyond RT-PCR cycles it is beloved to have low vial load. In this Study out of 3375 study participants 57%(n=1915) were positive with high viral load while rest of the 42% (n=1460) were found to be positive with low viral load as shown in the following chart:
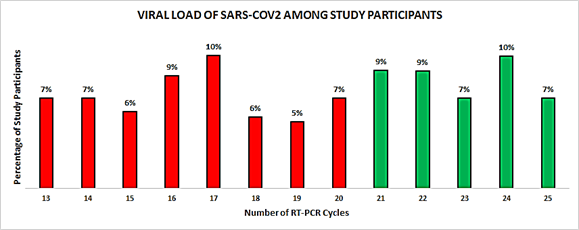
Chart 1: Viral load of study participants.
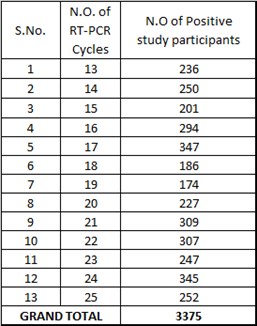
The following table further summarizes the above chart and is showing cycle wise positivity rate:
Table 1: Positivity rate against RT-PCR Cycles.
Frequency & Percentages of the Generalized/Non Specific Sign and Symptoms Among the Study Participants:
The following table summarizes the overall findings:
Table 2: Overall summary of the major findings.

Table 3: Table adopted from the above table.
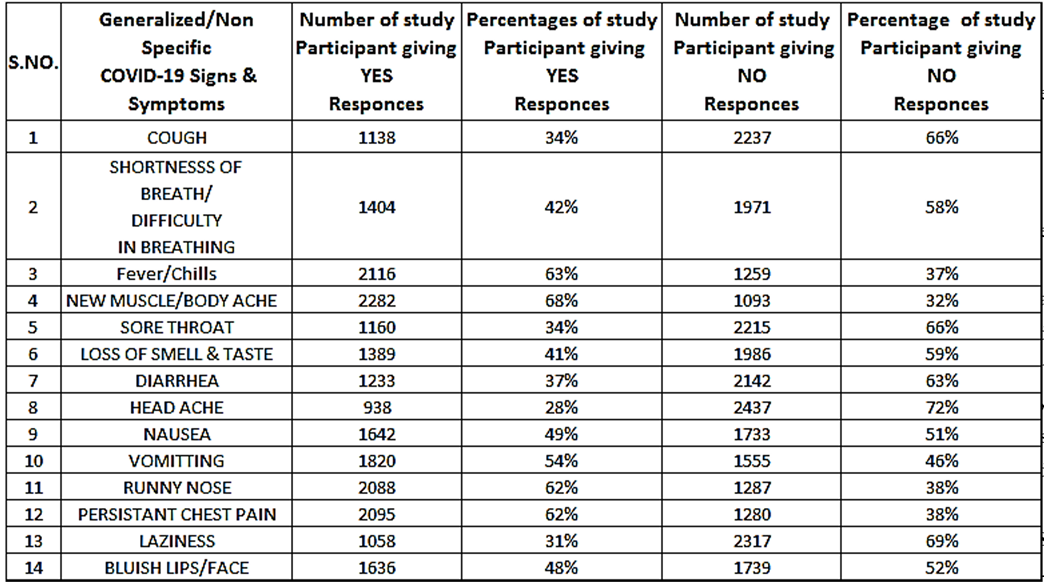
34% of the participants has cough while 66% had no cough, similarly 42% of the study participants reported to have S.O.B while 58% did not have any S.O.B. Moreover 63% had fever/chill while 37% did not had. New muscular or body ach was reported by 68% individuals while 32% had no new muscular/body ache. Similarly, sore throat was reported by 34% of the study participants while 66% of the study participants did not report any sore throat symptom.
Loss of smell and taste was reported by 41% of the study participants while 59% did not report this symptom. Diarrhea was reported by 37% of the study participants while 63% did not report any diarrhea. 28% of the study participants reported headache while 72% did not report any such symptom. Nausea was reported by 49% of the study participants while 51% did not report this symptom. Similarly, 54% of the study participants reported vomiting while 46% did not report this symptom. Runny nose was reported by 62% of the study participants while 38% did not report any such symptom. 68% of the study participants reported persistent chest pain while 38% did not report any chest pain. Laziness was reported by 31% of the study participants and lastly bluish lips/Face was reported by 48% of the study participants while 52% did not report any symptom.
Strength of association between generalized/non- specific covid-19 signs and symptoms reported by all study participants whose RT-PCR test was positive for all three SARS-COV2 genes (i.e., Orf + n + e) compare to others:
The following two tables summarizes the overall findings as shown below:
Table 4: Contingency 2x2 table between generalized/ non-specific COVID-19 Signs and symptoms reported by all the study participants whose RT-PCR report was positive for all the three SARS-COV 2 Genes (i.e., ORF + N + E) Compare to others.

Table 5: Odds Values, P-Values & C.I for table no 04.
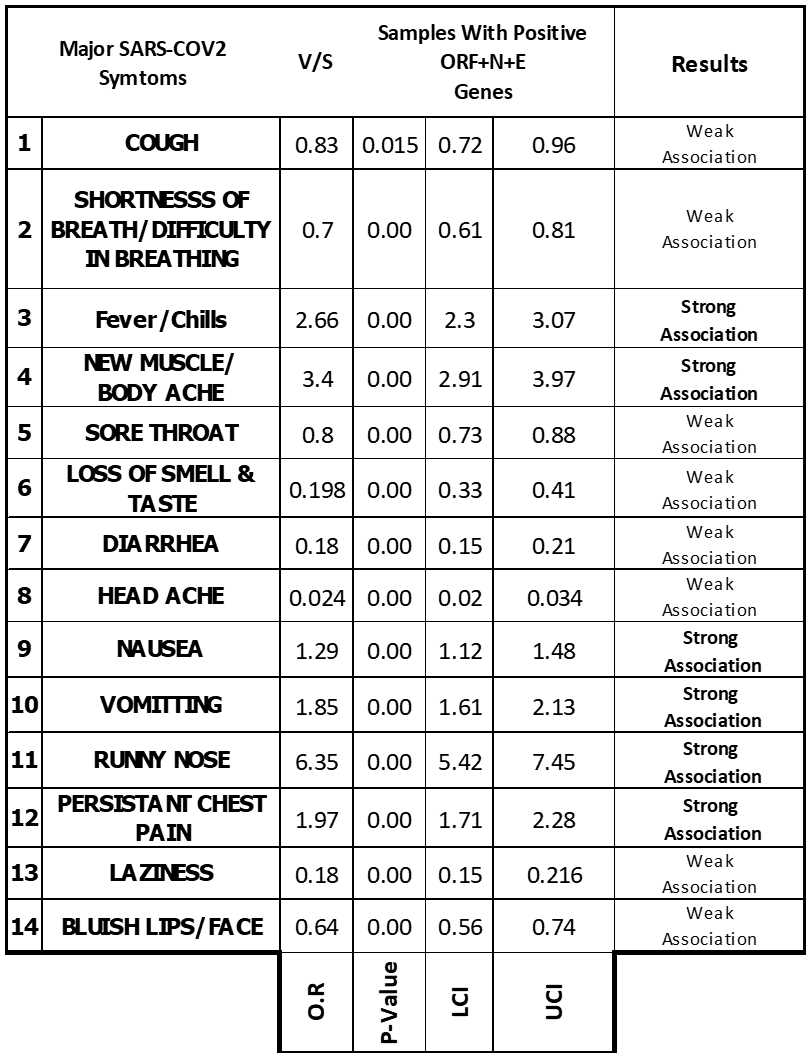
The presence of all three SARS-COV 2 genetic markers (i.e. ORF + N + E) was strongly associated with certain generalized/ nonspecific COVID-19 signs & symptoms like Fever/Chills, New Muscular/Bodily Ache, Nausea, Vomiting & Persistent chest pain.
Strength of association between generalized/non- specific covid-19 signs and symptoms reported by all study participants whose RT-PCR test was positive for different combinations of two SARS-COV2 genes (i.e., Orf, n, e) identified during rt-pcr process compare to others:
As previously mentioned during the RT-PCR analysis process among all the 3375 study participants only THREE different (ORF, N, E) paired combinations were identified as summarized below:
Table 6: Subclasses of Study participants with paired ORF, N, E genes Combinations.

Identified during RT-PCR:
Over all 3375 study participants were sub divided into three different classes the nasopharyngeal and throat samples of these study participant only yields a pair of 2 genes out of all the three major genes that is ORF, N, & E gene. One class of study participants were identified with only N & E genes. Similarly, one class of study participants were positive only with ORF & N gene while some of the study participants only express ORF & E genes on to their RT-PCR report. The details of each sub class along with their numbers and percentages are shown in the above table.
Table 7: Contingency 2x2 table between generalized/ non-specific COVID-19 Signs and symptoms reported by all the study participants whose RT-PCR report was positive for different combinations of two SARS-COV 2 Genes (i.e. ORF, N, E) Compare to others.


Strength of association between generalized/non- specific covid-19 signs and symptoms reported by all study participants whose RT-PCR test was positive for a single SARS-COV2 genes (i.e., Orf or n or e) compare to others:
Table 8: Contingency 2x2 table between generalized/ non-specific COVID-19 Signs and symptoms reported by all the study participants whose RT-PCR report was positive for a single SARS-COV 2 Genes (i.e. ORF/ N/ E) Compare to others.
The following two tables summarizes the overall findings as shown below:

Table 9: Odds Values, P-Values & C.I for table no 08.

Table 10: Odds Values, P-Values & C.I for table no 08.
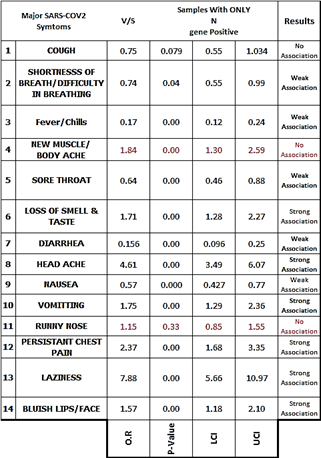
The presence of ORF gene was found to be strongly associated “ Shortness of Breath/Difficulty in Breathing”, Diarrhea, Head ache & Vomitting while Laiziness was found to have no association with the presence of ORF gene. Similarly the presence of N-gene was found to be strongly associated with Loss of smell & taste, Head ache,Presistant Chest Pain & Bluish lips/Face, N-gene was found to have no association with cough, New Muscular/Body ache & Runny Nose.
Table 11: Odds Values, P-Values & C.I for table no 08.
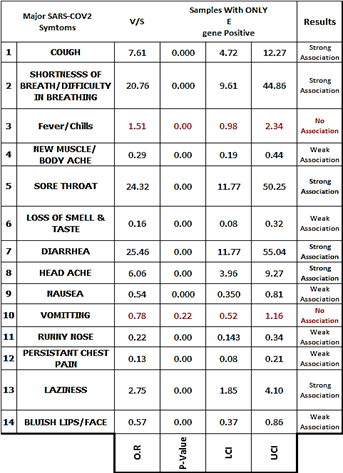
The presence of E-gene was strongly associated with Cough, Shortness of breath/ Difficulty in breathing, Sore throat, Diarrhea, Head ache & Laziness while the E-gene was found to have no association with Fever/Chill & Vomiting.
Discussion & Conclusion
Previously it has been well established from at least 23 different nations, with 26 different clinical presentations that Six symptoms—fever (58.66%), cough (54.52%), dyspnea (30.82%), malaise (29.75%), weariness (28.16%), and sputum/secretion (25.33%)—had a general prevalence more than or equal to 25%. Other prevalent symptoms included headache (12.17%), chest discomfort (11.49%), diarrhea (9.59%), sneezing (14.71%), sore throat (14.41%), rhinitis (14.29%), goosebumps (13.49%), dermatological signs (20.45%), anorexia (20.26%), myalgia (16.9%), and rhinitis. The manifestations of dermatology were only documented in one study. Hemoptysis was the least common indication or symptom (1.65%). The three most common symptoms in trials involving more than 100 patients were dyspnea (30.64%), cough (54.21%), and fever (57.93%) [1-13]. The study participants of our study also reported similar generalized/ nonspecific COVID-19 signs & symptoms.
From the very first case the SARS-COV 2 infection, a continuous mutation is reported in the virus as a result of which new variants popup frequently hence the overall virulence, treatment resistance, replication modalities & transmissions rates are all changing regularly hence in these circumstances it is of great importance to frequently monitor the symptomology of COVID-19. Currently no published literature has assessed the relationship of COVID-19 symptomology (i.e., COUGH, SHORTNESSS OF BREATH/DIFFICULTY IN BREATHING, Fever/Chills, NEW MUSCLE/BODY ACHE, SORE THROAT, LOSS OF SMELL & TASTE, DIARRHEA, HEAD ACHE, NAUSEA, VOMITTING, RUNNY NOSE, PERSISTANT CHEST PAIN, LAZINESS, BLUISH LIPS/FACE) with the ORF, N & E genes which are few of the diagnostic markers assessed for SARS COV-2 infection assessed during real-time PCR test.
The study findings showed that the presence of all three SARS-COV 2 genetic markers (i.e. ORF + N + E) was strongly associated with certain generalized/ nonspecific COVID-19 signs & symptoms like Fever/Chills, New Muscular/Bodily Ache, Nausea, Vomiting & Persistent chest pain.
Similarly, pair combination of different SARS-COV 2 virus genes identified showed that (ORF + E) gene pair presence was strongly associated Cough, Shortness of breath/Difficulty in breathing, New Muscular/Body pain, Sore throat, Runny nose and Bluish lips/face. Moreover, the (N + E) pair was found to be strongly associated with Loss of smell & Taste, Head ache, Vomiting & Persistent chest pain/pressure. Furthermore, the (ORF + N) pairing was found to be strongly associated with Diarrhea, Nausea & Laziness.
Our study also showed that individually the presence of ORF gene was found to be strongly associated “ Shortness of Breath/Difficulty in Breathing”, Diarrhea, Head ache & Vomitting while Laiziness was found to have no association with the presence of ORF gene. Similarly the presence of N-gene was found to be strongly associated with Loss of smell & taste, Head ache,Presistant Chest Pain & Bluish lips/Face, N-gene was found to have no association with cough, New Muscular/Body ache & Runny Nose. While the presence of E-gene was strongly associated with Cough, Shortness of breath/ Difficulty in breathing, Sore throat, Diarrhea, Head ache & Laziness while the E-gene was found to have no association with Fever/Chill & Vomiting.
References
- da Rosa Mesquita R, Francelino Silva Junior LC, Santos Santana FM, Farias de Oliveira T, Campos Alcântara R, Monteiro Arnozo G, et al. Clinical manifestations of COVID-19 in the general population: systematic review. Wiener klinische Wochenschrift, 2021; 133(7-8): 377–382. https://doi.org/10.1007/s00508-020-01760-4.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020; 579(7798): 1–4. doi: 10.1038/s41586-020-2012-7.
- Tu H, Tu S, Gao S, Shao A, Sheng J. The epidemiological and clinical features of COVID-19 and lessons from this global infectious public health event. J Infect, doi: 10.1016/j.jinf.2020.04.011.
- Center for Disease Control and Prevention. Coronavirus disease (COVID-19), 2020.
- Liu Y, Mao B, Liang S, Yang JW, Lu HW, Chai YH, et al. Association between age and clinical characteristics and outcomes of COVID-19. European Respiratory Journal, 2020; 55(5).
- Wu Z, McGoogan JM. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA, 2020; 323(13): 1239–1242. https://doi.org/10.1001/jama.2020.2648
- Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses, 2019; 11(1): 59. https://doi.org/10.3390/v11010059
- Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell host & microbe, 2020; 27(3): 325–328. https://doi.org/10.1016/j.chom.2020.02.001
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England), 2020; 395(10224): 565–574. https://doi.org/10.1016/S0140-6736(20)30251-8
- Guruprasad K. Geographical Distribution of Amino Acid Mutations in Human SARS-CoV-2 Orf1ab Poly-Proteins Compared to the Equivalent Reference Proteins from China. ChemRxiv, 2021. doi:10.26434/chemrxiv-2021-lf2zd-v2
- Chang TJ, Yang DM, Wang ML, Liang KH, Tsai PH, Chiou SH, et al. Genomic analysis and comparative multiple sequences of SARS-CoV2. Journal of the Chinese Medical Association: JCMA, 2020; 83(6): 537–543. https://doi.org/10.1097/JCMA.0000000000000335
- Astuti I, Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes & metabolic syndrome, 2020; 14(4): 407–412. https://doi.org/10.1016/j.dsx.2020.04.020
- Su S, Wong G, Shi W, Liu J, Lai A, Zhou J, et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends in microbiology, 2016; 24(6): 490–502. https://doi.org/10.1016/j.tim.2016.03.003
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annual review of microbiology, 1997; 51: 151–178. https://doi.org/10.1146/annurev.micro.51.1.151
- Holmes EC, Rambaut A. Viral evolution and the emergence of SARS coronavirus. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 2004; 359(1447): 1059–1065. https://doi.org/10.1098/rstb.2004.1478
- Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene reports, 2020; 19: 100682. https://doi.org/10.1016/j.genrep.2020.100682
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases, 2020; 81: 104260. https://doi.org/10.1016/j.meegid.2020.104260
- Yu WB, Tang GD, Zhang L, Corlett RT. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2 / HCoV-19) using whole genomic data. Zoological research, 2020; 41(3): 247–257. https://doi.org/10.24272/j.issn.2095-8137.2020.022
- Holland LA, Kaelin EA, Maqsood R, Estifanos B, Wu LI, Varsani A, et al. An 81-Nucleotide Deletion in SARS-CoV-2 ORF7a Identified from Sentinel Surveillance in Arizona (January to March 2020). Journal of virology, 2020; 94(14): e00711-20. https://doi.org/10.1128/JVI.00711-20
- Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Scientific reports, 2018; 8(1): 15177. https://doi.org/10.1038/s41598-018-33487-8
- Su Y, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, et al. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio, 2020; 11(4): e01610-20. https://doi.org/10.1128/mBio.01610-20
- Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. Journal of human genetics, 2020; 65(12): 1075–1082. https://doi.org/10.1038/s10038-020-0808-9
- Ahamad S, Kanipakam H, Gupta D. Insights into the structural and dynamical changes of spike glycoprotein mutations associated with SARS-CoV-2 host receptor binding. Journal of biomolecular structure & dynamics, 2022; 40(1): 263–275. https://doi.org/10.1080/07391102.2020.1811774
- Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell, 2020; 182(5): 1284–1294.e9. https://doi.org/10.1016/j.cell.2020.07.012
- Benvenuto D, Angeletti S, Giovanetti M, Bianchi M, Pascarella S, Cauda R, et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. The Journal of infection, 2020; 81(1): e24–e27. https://doi.org/10.1016/j.jinf.2020.03.058

