Analysis of EGFR Mutation by Rapid PCR Methods Can Yield Better Quality Results Using Intra-Operative Frozen Section Tissue In Early Stage, Non-Small Cell Lung Cancer Patients
Alison Finall1,*, Kate Hurlow1, Kate Murphy1, Gareth Leopold1, Tawfik Elazzabi1, Ira Goldsmith2 and Suprotim Basu3
1Cellular and Molecular Pathology, Morriston Hospital, Swansea Bay University Health Board, Swansea UK
2Department of Cardiothoracic Surgery, Morriston Hospital, Swansea Bay University Health Board, Swansea UK
3Department of Oncology, Morriston Hospital, Swansea Bay University Health Board, Swansea UK
Received Date: 17/02/2023; Published Date: 13/04/2023
*Corresponding author: Dr. Alison Finall, Consultant Histopathologist, Cellular and Molecular Pathology, Morriston Hospital, Swansea SA6 6NL, Swansea Bay University Health Board, Swansea UK
Abstract
Introduction: Osimertinib is now licensed for use in early-stage non-small cell lung cancer patients (NSCLC) as an adjunct in the post-surgical setting. Identification of appropriate patients for this therapy relies upon identification of somatic epidermal growth factor receptor (EGFR) mutations. The IdyllaTM EGFR Mutation Test is a rapid, fully automated near patient test that could be used in histopathology laboratories to identify such patients.
Methods: We conducted a pilot study of 12 consecutive patients with non-squamous NSCLC histology undergoing intra-operative frozen section diagnosis over a two-year period to determine the suitability of frozen tissue for EGFR testing as compared with matched formalin-fixed paraffin-embedded (FFPE) tissue samples.
Results: There was a 100% concordance of findings between the IdyllaTM EGFR Mutation tests conducted on frozen section and FFPE tissue samples. There was also full concordance with next generation sequencing (NGS) results, where performed. The cycling quotient (CQ) value for fresh frozen tissue samples was significantly lower than that for FFPE samples (p<0.0001).
Conclusions: This is the first study to assess suitability of DNA in fresh frozen samples at time of intra-operative frozen section for NSCLC patients using a rapid, automated, singe gene polymerase chain reaction (PCR) method. This test could be used to identify appropriate patients for post-surgical, adjuvant Osimertinib therapy. There are potential cost and time savings by choosing this singe gene test rather than utilising NGS methods for early-stage NSCLC patients.
Introduction
Tyrosine kinase inhibitors have been used in the treatment of patients with disseminated Non-Small Cell Lung Cancer (NSCLC) harbouring epidermal growth factor receptor (EGFR) mutations for over ten years [1-4]. Mutations in the somatic lung EGFR gene drive cells to divide and proliferate and result in adenocarcinomas in 10- 20% of patients in the UK [5]. Osimertinib is a third-generation tyrosine kinase inhibitor drug with irreversible binding affinity for the EGFR receptor that can give patients extended survival in NSCLC [6]. It has a better safety profile than standard, platinum-based chemotherapy and has the added benefit of administration as an oral agent in the community [6]. However, standard care for early-stage NSCLC patients (stage IB, II or IIIA) is surgical resection by lobectomy and regional lymph node dissection and this mode of management applies to a third of lung cancer patients overall [6]. Unfortunately, up to three quarters of patients will develop disease recurrence even with post-operative standard chemotherapy [7]. The ADUARA trial showed that patients with early-stage NSCLC receiving adjuvant Osimertinib post-surgery had a longer cancer free survival interval compared with controls where patients had somatic EGFR mutations in exons 19 and 21 of their tumour [8]. The Medicines and Healthcare products Regulatory Agency (MHRA) have recently licensed Osimertinib for use in the UK for patients with early stage, EGFR-mutated NSCLC in adults following complete surgical resection on the basis of this evidence [9].
We sought to determine whether a fully-automated, rapid PCR assay could be used at the time of intraoperative frozen section in NSCLC patients to support this medical advance and to help surgical oncologists identify suitable patients for personalised adjuvant therapy in a short-time frame. Some patients with a lung lesion identified on CT are not eligible for pre-operative diagnosis due to inaccessibility or high-risk of serious morbidity as a consequence of image guided core biopsy. These patients have an intra-operative frozen section to confirm malignancy at the start of the resection surgery before proceeding with a lobectomy and nodal dissection.
We compared CQ values as a measure of DNA quality input into the fully automated rapid PCR console with paired formalin fixed, paraffin embedded tissue samples from the same patients. Previously, we have verified the IdyllaTM EGFR Mutation Test (a rapid automated PCR assay) as a suitable adjunct to biomarker detection pathways by comparing with next generation sequencing [10]. We found a detection concordance of 96.3 % for detection of EGFR point mutations in DNA between the two assay methods [10]. The rapid PCR method offers the possibility for same day detection of the most common EGFR variants in NSCLC which is particularly important for patients with stage 4 disease who are at risk of rapid clinical deterioration, as acknowledged in the NHS England salvage pathway [11].
Methods
Case selection
Consecutive patients undergoing intra-operative frozen section diagnosis during surgical resection of a lung mass were potentially eligible for inclusion in the study. Patients underwent the IdyllaTM EGFR Mutation Test following standard of care procedures for frozen section only after identification of morphology to suggest a diagnosis of carcinoma, NOS or adenocarcinoma favouring lung primary. Patients with factors suspicious for a diagnosis of tuberculosis are not eligible for frozen section diagnosis. Minimum sample requirements for rapid automated testing included 10% tumour nuclear content (TNC) and the ability to take a 5-10mm thick tissue section. Samples were frozen to -40oC using the automated PrestoCHILL (Milestine Medical) platform with MMC, MilestoneTM Cryoembedding Compound.
An adjacent fresh tissue sample was selected for processing into a Formalin Fixed Paraffin Embedded (FFPE) tissue block for quality assessment of material submitted rapid PCR. The test used fresh and FFPE paired samples left over following standard of care requirements.
Case mix
All patients with non-squamous, non-small cell neuroendocrine carcinoma of the lung identified by intra-operative frozen section histopathological analysis were included in the study on a non-selected, all-comers basis. Palpation guided intra-operative biopsy or wedge resection of lung or peripheral tumours were equally eligible for inclusion. Patients under the age of 18 years were not included.
Pre-analytical considerations
Sufficient fresh non-squamous, NSCLC tumour samples were required for testing as described above. A second block of tissue was selected from the opposing face of the dissected fresh sample for processing into an FFPE block. FFPE processing required fixation in 10% neutral buffered formalin for at least 6 hours prior to automated, standard overnight protocol processing on Thermo Fisher Scientific Exselsior ES tissue processors (2008). The oldest FFPE block used in the study dates from the end February 2020. Testing concluded by mid-September 2022. The maximum storage time for FFPE tissue blocks was 31 months.
Idylla EGFR Mutation Test
Tissue sections, frozen and FFPE, were taken from the microtome direct to the ‘IdyllaTM EGFR Mutation Test’ cartridge directly without use of a water bath or mounting on glass slides. Due to the nature of the procedure and sample size, microdissection was not required for TNC enrichment. Frozen and allied FFPE tissue slides were assessed for TNC by a single pathologist (AF) according to published guidance and previous work [12,13]. Tissue samples were sandwiched between moist filter paper discs to ensure adherence to test platform in the cartridge base. Molecular grade, nuclease-free water was used to moisten the filter paper discs. Automated DNA extraction within the IdyllaTM EGFR Mutation test cartridge prevents sample contamination and need for molecular grade facilities within the histopathology department. Pre-determined primer sets identify the presence of up to 51 specific EGFR mutations within rapid, automated assay by detecting fluorescence above a proprietary threshold during PCR. Cycling Quotient (CQ) values were recorded as a proxy measure of DNA quality where lower CQ values indicate fewer cycles of PCR being required to detect control fluorescence. A CQ value of 20 is equivalent to 200ng of DNA as determined in experiments conducted by the manufacturer [10,14]. A CQ value of 19.8 was generated for EGFR control samples containing 2500 copies of the gene and 21.3 for 1000WT copies of EGFR control gene input [14]. The rapid PCR assay has a CE-IVD certificate from the MRHA for use as a clinical diagnostic assay based on FFPE tissue samples [14]. The Limit of detection of the test is less than 5% TNC for mutations in exons 19, 20 and 21 of the EGFR gene and <10% for exon 18 [14]. Only tumour exon 19 deletions and the L858R mutation in exon 21 of the EGFR gene are relevant to the licensed prescription of Osimertinib in the clinical context of post-surgical adjuvant treatment [9]. The ‘IdyllaTM EGFR Mutation Test’ cartridge contains PCR primers to detect both of these common EGFR mutations that occur in a minority of NSCLC patients. This study was designed to determined suitability of fresh frozen tissue as a potential DNA input source for rapid reporting of EGFR mutations where clinically appropriate.
Ethical considerations
The study proposal was reviewed the Swansea Bay University Health Board Joint Study Review Committee and deemed service development. Audit, service development and quality improvement projects are exempt from the need for research ethics review [15,16]. No randomisation or management/treatment interventions took place as part of, or consequence of, this project. The findings may be of use to our local patient cohort and are not generalisable. Compliance with Human Tissue Authority guidance on use of diagnostic human tissue is assured. Verification of all new assays require in-house verification to meet ISO15189 medical diagnostic laboratory standards.
Statistical analysis
Descriptive statistics and a paired t-test were prepared using SPSS V.26.0.0 statistical package from IBM and graphad.com. Study was closed when statistical significance for differences between CQ values between the paired sample types was achieved. CQ values were assumed to follow a normal distribution.
Results
Twelve patients underwent frozen section with non-squamous, Non-Small Cell Neuroendocrine Carcinoma (NSCLC) morphology between February 2020 and May 2022. One of these frozen section samples was compromised and could not be tested as a fresh frozen sample leaving eleven cases suitable for analysis. All samples submitted were peripheral lung wedge resections for primary frozen section diagnosis.
Table 1: Clinical background, staging, treatment and follow-up status of patients undergoing intra-operative thoracic frozen section.
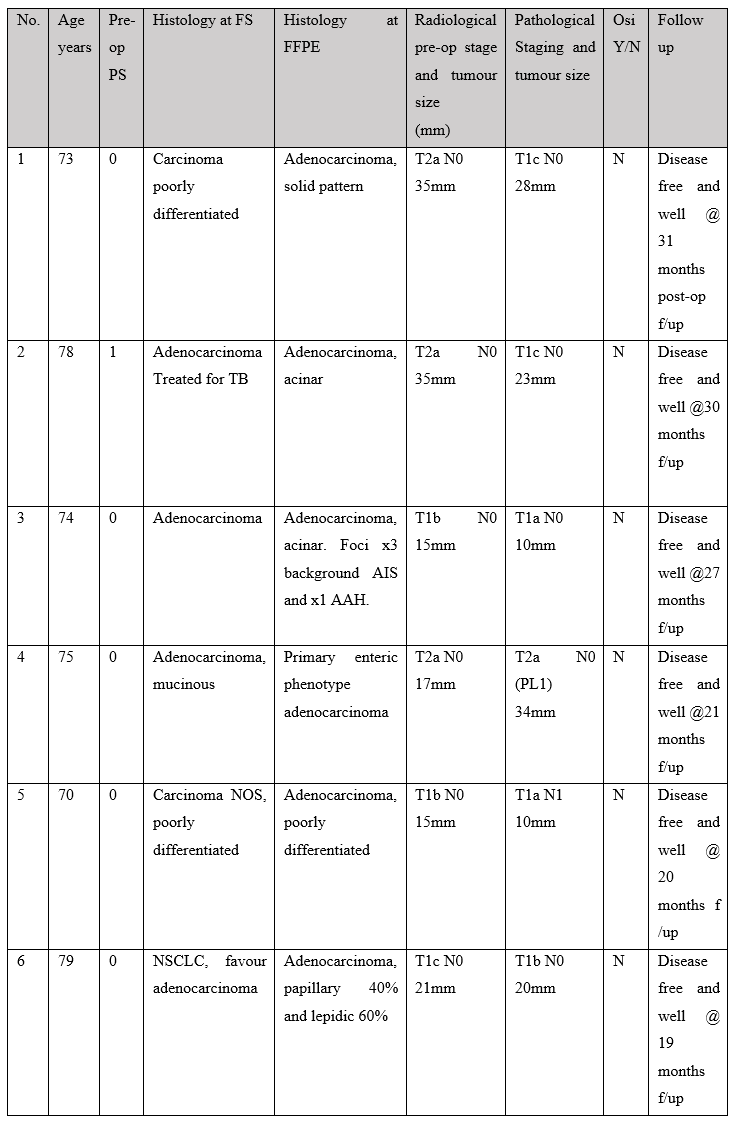
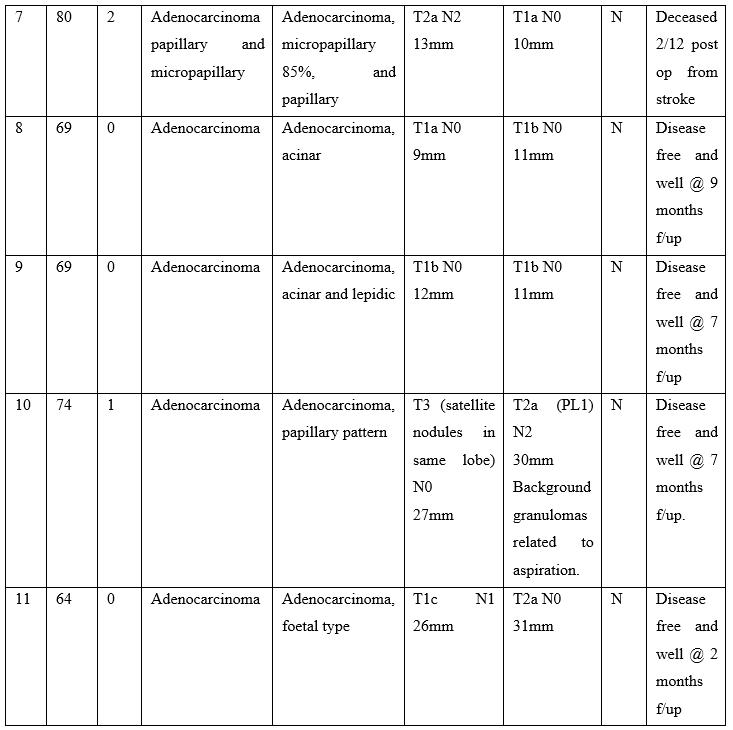
Table 1 Abbreviations: T, tumour; N, lymph node; PL, pleural invasion; EGFR, epidermal growth factor receptor; TPS, performance status; FS, frozen section; FFPE, formalin fixed, paraffin embedded; Osi, Osimertinib; No, case number; f/up, follow-up; TB, Mycobacterium tuberculosis.
Most patients in the cohort had a performance status of 0 and a successful outcome following surgery. One patient was deceased 2 months after surgery due to cerebral infarction. See Table 1 for clinical details. None of our patients received adjuvant Osimertinib in the post-operative period. One patient started adjuvant Carboplatin treatment in a setting of regional lymph node metastases being identified in the surgical resection specimen. Two cases were reported as carcinoma, not otherwise specified at frozen section; Both of these cases were poorly differentiated adenocarcinoma as determined by immunohistochemistry performed on the formalin fixed, paraffin embedded (FFPE) tumour blocks.
The Cycling Quotient (CQ) value is a proxy reflection of the amount and quality of DNA available or polymerase chain reaction (PCR) in the automated EGFR mutational test. A low CQ value indicates that fewer cycles of PCR were required for sufficient levels of florescence to be detected for a valid call by the IdyllaTM instrument software. The instructions for use document for IdyllaTM EGFR Mutation Test, states an optimal CQ value range of 19 and 24 for clinical reliability of use. The difference between the CQ values between the groups was determined using the paired T-test as both groups of CQ values were from the same patient sample and therefore considered dependent. See table 2. The two-tailed p-value is less than 0.0001, which indicates an extremely statistically significant difference.
Table 2: EGFR analysis using a rapid PCR assay and fresh frozen or formalin fixed paraffin embedded (FFPE) tissue.

Abbreviations: EGFR, epidermal growth factor receptor; TAT, turn-around time; FS, frozen section; FFPE, formalin fixed, paraffin embedded; TNC, tumour nuclear content, CQ, cycling quotient; ND, not done; WT, wild-type; NR, not recorded.
All eleven samples were devoid of EGFR mutation by rapid PCR and next generation sequencing (NGS). The outcome of the rapid PCR test was the same for FFPE samples and fresh sections samples in 100% of cases. The study commenced before Osimertinib was approved for clinical use in a post-surgical adjuvant setting in early-stage NSCLC. Until October 2021, pathologists in our health board only reflex requested DNA NGS for those patients with stage 3 and stage 4 disease in an effort to target valuable resources to cases of clinical utility.
Table 3: Statistical Data from Paired T-test.
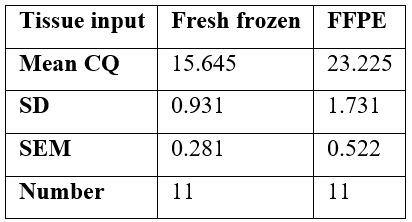
Abbreviations: CQ, Cycling Quotient; FFPE, formalin fixed, paraffin embedded; SD, standard deviation; SEM, Standard error of the mean.
Discussion
Our study is the first to assess frozen tissue for molecular analysis of NSCLC tissue during intra-operative frozen section diagnosis. Intra-operative molecular analysis has been trialled in breast cancer patient to refine diagnosis of metastatic disease within axillary sentinel lymph nodes with good results but a role for molecular diagnostics has yet to be demonstrated in other clinical settings [17,18]. Some rapid, fully-automated PCR assays by Biocartis are designed specifically for use with fresh and frozen samples in the context of infectious disease but their solid tumour oncology assays are designed for use with FFPE tissue [19,20]. A paper by Dagogo-Jack et al, demonstrated that the Idylla EGFR assay could be used with frozen tissue input in a workflow designed to make ultra-rapid diagnostic information available to guide oncologic treatment in an advanced disease setting [21]. Some authors have also verified use of IdyllaTM cartridges with pre-extracted DNA and using fluid cytology specimens [22-24].
Our data suggest that the quality and quantity of DNA available for PCR in the fully automated, rapid PCR assay was greater in the fresh frozen tissue samples than in the subsequent matched FFPE tissue samples. This is an expected conclusion as formalin-induced protein-DNA cross linking requires harsh chemical treatment to liberate DNA for PCR. The process of DNA extraction from FFPE tissue results in single strand breaks and smaller nucleotide length, as would be reflected in the CQ value [25]. Despite this the rapid, fully-automated PCR assay was designed for use in lung cancer specimens in standard histopathology practice and based on FFPE tissue validation protocols. As such, the instructions for use indicate a recommended CQ spectrum of between 18 and 25 for wild-type EGFR DNA [14]. The mean CQ value found for FFPE was 23.225 and for fresh frozen tissue was 15.645 in our paired samples set and was statistically significant (p>0.0001). FFPE results were well within the expected CQ range. For the fresh samples, technically, these are outside the expected range for the test as it is designed for use but suggests significant improved yields of DNA for PCR which is likely because of less formaldehyde induced damage. Our findings appear to be at odds with Sorber et al, however, who describe deterioration of nucleotide content compared with paired FFPE lung malignancy samples during their verification of the IdyllaTM GeneFusion assay [18]. However, the Sorber study focussed on RNA content for the IdyllaTM GeneFusion assay and used frozen tissue samples that had been stored for up to 9 years at -80oC prior to analysis [26]. It is well known that RNA is a much more fragile nucleotide molecule in comparison to DNA and can suffer degradation from nuclease enzymes in the environment and are more sensitive to hydrolysis because of the additional hydroxyl group that forms part of the ribose structure [27]. Most studies agree that the quantity and quality of extractable DNA is superior in fresh frozen tissue compared with FFPE archived tissue [17,18,28,29, 30-32] and this is clearly reflected in our data also.
In our centre, we usually perform in the region of 75 frozen sections per annum (1 or 2 per week). This number was substantially reduced by restrictions to operative practices during the pandemic and is a limitation of our study. Indeed, frozen sections diagnosis was actively discouraged early in the pandemic as the procedure generates aerosol droplets containing microscopic particulate matter and putting biomedical scientist staff at risk of developing COVID-19. This study commenced early during the COVID-19 pandemic and a short fall of operative procedures during this time gave limited numbers of intra-operative frozen section samples for analysis. All of the submitted samples were to determine whether the radiologically-identified lung mass was benign or malignant in order to influence immediate surgical management of the patient. In our centre, the majority of patients have their surgical treatment planned on the basis of a pre-operative diagnostic biopsy, which may be bronchial, CT-guided or from endobronchial ultrasound-guided (EBUS) fine needle aspiration cytology of mediastinal lymph nodes. In a minority of patients, the tumour may be inaccessible to a biopsy needle or endoscope or biopsy may be deemed as too high-risk due to anatomical proximity to essential structures and/or pre-existing co-morbidities that severely restrict lung function. A further limitation of our work is not having direct access to molecular laboratory facilities to extract DNA from the paired fresh and FFPE tissue samples and attempt to quantify the DNA content of each, e.g., using a Qubit Fluorometer or NanoDrop spectrophotometer (Thermo Fisher ScientificTM) [33].
At the time of writing, Osimertinib is the only targeted therapy indicated in the post-surgical setting as an adjuvant treatment in early-stage NSCLC [8,9,34,35]. The decision to treat with adjunct Osimertinib relies on information from the EGFR gene alone and so this represents an indication for single gene testing by PCR. This approach could save considerable amounts of money for the NHS, and in other healthcare settings, by refraining from a blanket approach of DNA NGS sequencing for all lung cancer patients. It may be more in keeping with principles of prudent healthcare to choose this single gene test at around £150 per sample for the rapid automated PCR test as opposed to £785 (Illumina TrusightTM NGS panel) per sample cost [36]. At a time of rising inflation and NHS resource constraints, the issue of cost cannot be ignored particularly when early-stage NSCLC patients make up around 30% of patient cohorts. In addition to the cost savings, the turnaround time for individual tests is 180 minutes with minimal (2 minutes) of operator time and suitability for use in histopathology laboratories without molecular grade facilities needed for nucleic acid extraction. We have previously shown that turnaround time is of critical importance for late-stage NSCLC patients [10]. Though this may not be of such clinical concern in the adjuvant setting, the cost and efficiency savings are attractive.
Take home lessons:
- This is the first study to assess suitability of DNA in fresh frozen samples at time of intra-operative frozen section for NSCLC patients using the IdyllaTM EGFR Mutation Test.
- DNA quantity and quality is significantly better using fresh frozen samples compared with paired FFPE samples as reflected in the CQ value (p<0.0001).
- Fully automated, rapid PCR could be used to identify appropriate patients for post-surgical, adjuvant Osimertinib therapy in surgical pathology reporting.
- There are potential cost and time savings by choosing this singe gene test rather than utilising NGS methods for early-stage NSCLC patients.
References
- Mok TS, Zhou Q, Leung L, Loong HH. Personalized medicine for non-small-cell lung cancer. Expert Rev Anticancer Ther, 2010; 10(10): 1601-1611.
- Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol, 2015; 16(2): 141-151.
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med, 2009; 361(10): 947-957.
- Mok TSK, Kim SW, Wu YL, Nakagawa K, Yang JJ, Ahn MJ, et al. Gefitinib Plus Chemotherapy Versus Chemotherapy in Epidermal Growth Factor Receptor Mutation-Positive Non-Small-Cell Lung Cancer Resistant to First-Line Gefitinib (IMPRESS): Overall Survival and Biomarker Analyses. J Clin Oncol, 2017; 35(36): 4027-4034.
- Graham RP, Treece AL, Lindeman NI, Vasalos P, Shan M, Jennings LJ, et al. Worldwide Frequency of Commonly Detected EGFR Mutations. Arch Pathol Lab Med, 2018; 142(2): 163-167.
- Lee CS, Milone M, Seetharamu N. Osimertinib in EGFR-Mutated Lung Cancer: A Review of the Existing and Emerging Clinical Data. Onco Targets Ther, 2021; 14: 4579-4597.
- Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in Resected. N Engl J Med, 2020; 383(18): 1711-1723.
- Wu YL, Herbst RS, Mann H, Rukazenkov Y, Marotti M, Tsuboi M. ADAURA: Phase III, Double-blind, Randomized Study of Osimertinib Versus Placebo in EGFR Mutation-positive Early-stage NSCLC After Complete Surgical Resection. Clin Lung Cancer, 2018; 19(4): e533-e536.
- NICE. Osimertinib for adjuvant treatment of EGFR mutation positive NSCLC in adults after complete tumour resection.
- Finall A, Davies G, Jones T, Emlyn G, Huey P, Mullard A. Integration of rapid PCR testing as an adjunct to NGS in diagnostic pathology services within the UK: evidence from a case series of non-squamous, non-small cell lung cancer (NSCLC) patients with follow-up. J Clin Pathol, 2022.
- Group NGSTW. Solid Cancer Genomics Testing: The Salvage Pathway. London: NHS England and NHS Improvement, 2021.
- Dufraing K, van Krieken JH, De Hertogh G, Hoefler G, Oniscu A, Kuhlmann TP, et al. Neoplastic cell percentage estimation in tissue samples for molecular oncology: recommendations from a modified Delphi study. Histopathology, 2019; 75(3): 312-319.
- Finall A, Murphy K, Frazer RD. Improving care of melanoma patients through efficient, integrated cellular-molecular pathology workflows using tissue samples with low tumour nuclear content. J Clin Pathol, 2022.
- Biocartis. Instructions for use: Idylla EGFR Mutation Test. Biocartis NV; 2017. p. 1-41.
- NRES. Defining Research. In: Agency NPS, editor.
- Casarett D, Karlawish JH, Sugarman J. Determining when quality improvement initiatives should be considered research: proposed criteria and potential implications. JAMA, 2000; 283(17): 2275-2280.
- Norton N, Sun Z, Asmann YW, Serie DJ, Necela BM, Bhagwate A, et al. Gene expression, single nucleotide variant and fusion transcript discovery in archival material from breast tumors. PLoS One, 2013; 8(11): e81925.
- Bhagwate AV, Liu Y, Winham SJ, McDonough SJ, Stallings-Mann ML, Heinzen EP, et al. Bioinformatics and DNA-extraction strategies to reliably detect genetic variants from FFPE breast tissue samples. BMC Genomics, 2019; 20(1): 689.
- Matheeussen V, Loens K, Kuijstermans M, Jacobs K, Coenen S, van der Velden AW, et al. Diagnostic performance of the Idylla™ respiratory panel for molecular detection of influenza A/B in patients presenting to primary care with influenza-like illness during 3 consecutive influenza seasons. J Clin Virol, 2021; 144: 104998.
- Wouters Y, Keyaerts E, Rector A, Van Even E, Vissers S, Koletzki D, et al. Comparison of the Idylla™ Respiratory (IFV-RSV) panel with the GeneXpert Xpert® Flu/RSV assay: a retrospective study with nasopharyngeal and midturbinate samples. Diagn Microbiol Infect Dis, 2019; 94(1): 33-37.
- Dagogo-Jack I, Azzolli CG, Fintelmann F, Mino-Kenudson M, Farago AF, Gainor JF, et al. Clinical Utility of Rapid EGFR Genotyping in Advanced Lung Cancer. JCO Precis Oncol, 2018; 2018.
- Grant J, Stanley A, Balbi K, Gerrard G, Bennett P. Performance evaluation of the Biocartis Idylla EGFR Mutation Test using pre-extracted DNA from a cohort of highly characterised mutation positive samples. J Clin Pathol, 2021.
- De Luca C, Vigliar E, d'Anna M, Pisapia P, Bellevicine C, Malapelle U, et al. detection on archival cytological smears by the novel fully automated polymerase chain reaction-based Idylla mutation test. Cytojournal, 2017; 14: 5.
- de Biase D, de Luca C, Gragnano G, Visani M, Bellevicine C, Malapelle U, et al. Fully automated PCR detection of KRAS mutations on pancreatic endoscopic ultrasound fine-needle aspirates. J Clin Pathol, 2016.
- Grafstrom RC, Fornace AJ, Autrup H, Lechner JF, Harris CC. Formaldehyde damage to DNA and inhibition of DNA repair in human bronchial cells. Science, 1983; 220(4593): 216-218.
- Sorber L, Van Dorst B, Bellon E, Zwaenepoel K, Lambin S, De Winne K, et al. NTRK Gene Fusion Detection in a Pan-Cancer Setting Using the Idylla GeneFusion Assay. J Mol Diagn, 2022; 24(7): 750-759.
- Elliott D, Ladomery M. Molecular biology of RNA. Oxford: Oxford University Press, 2011.
- Gao XH, Li J, Gong HF, Yu GY, Liu P, Hao LQ, et al. Comparison of Fresh Frozen Tissue With Formalin-Fixed Paraffin-Embedded Tissue for Mutation Analysis Using a Multi-Gene Panel in Patients With Colorectal Cancer. Front Oncol, 2020; 10: 310.
- Mullegama SV, Alberti MO, Au C, Li Y, Toy T, Tomasian V, et al. Nucleic Acid Extraction from Human Biological Samples. Methods Mol Biol, 2019; 1897: 359-383.
- Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One, 2014; 9(5): e98187.
- Wang JH, Gouda-Vossos A, Dzamko N, Halliday G, Huang Y. DNA extraction from fresh-frozen and formalin-fixed, paraffin-embedded human brain tissue. Neurosci Bull, 2013; 29(5): 649-654.
- McDonough SJ, Bhagwate A, Sun Z, Wang C, Zschunke M, Gorman JA, et al. Use of FFPE-derived DNA in next generation sequencing: DNA extraction methods. PLoS One, 2019; 14(4): e0211400.
- Bapat PR, Epari S, Joshi PV, Dhanavade DS, Rumde RH, Gurav MY, et al. Comparative Assessment of DNA Extraction Techniques From Formalin-Fixed, Paraffin-Embedded Tumor Specimens and Their Impact on Downstream Analysis. Am J Clin Pathol, 2022.
- Jones DR, Wu YL, Tsuboi M, Herbst RS. Targeted therapies for resectable lung adenocarcinoma: ADAURA opens for thoracic oncologic surgeons. J Thorac Cardiovasc Surg, 2021; 162(1): 288-292.
- Wu YL, John T, Grohe C, Majem M, Goldman JW, Kim SW, et al. Postoperative Chemotherapy Use and Outcomes From ADAURA: Osimertinib as Adjuvant Therapy for Resected EGFR-Mutated NSCLC. J Thorac Oncol, 2022; 17(3): 423-433.
- Kumar S, Bennett A, Campbell PA, Palidwor G, Lo B, Perkins TJ, et al. Costs of Next-Generation Sequencing Assays in Non-Small Cell Lung Cancer: A Micro-Costing Study. Curr Oncol, 2022; 29(8): 5238-5246.

