Comparing the Results from Faecal Microbiome Transplantation (Fmt) and Gut Flora Replacement Therapy (Gfrt) using 16s Mrna Microbiome Mapping
Julian N. Kenyon*
The Dove Clinic, Twyford, Winchester, England
Received Date: 16/06/2021; Published Date: 06/07/2021
*Corresponding author: Julian N Kenyon, The Dove Clinic, Twyford, Winchester, Hampshire, SO21 1NT, England. Telephone: 01962 718000, 07740786342 (Mobile); Email: jnkenyon@doveclinic.com
Abstract
We compared the results from 10 randomly chosen FMT patients and 10 randomly chosen patients using GFRT. We used 16S mRNA microbiome mapping on all of these patients. We identified that GFRT is just as effective as FMT, a great deal safer and is a necessary step to protect patients from catching COVID-19.
Keywords: Faecal Microbiome Transplantation (FMT); Gut Flora Replacement Therapy (GFRT); COVID-19; 16s mRNA microbiome mapping
Introduction
We have previously used FMT, this is well researched [1] and there has been significant success in difficult to treat conditions [2].
With the advent of the pandemic, we had to reconsider our use of donor derived laboratory processed implants because of the potential transmission of COVID-19 from exposure to such implants [3-5]
What we also noted is that the now standard use of measurements in sewage water waste by water authorities, measuring the levels of COVID-19 in the sewage effluent has proven to be a reliable indicator of the prevalence of local outbreaks and indeed, this level of COVID-19 in the sewage effluent predates the onset of an outbreak of COVID-19 locally [6].
We have seen many claims of patients catching COVID-19 from the stool of COVID-19 sufferers, these notifications seem to be increasingly common, but in practice they are not provable. The presence of large amounts of COVID-19 in faecal matter raises the possibility that part of the explanation as to why COVID-19 is so contagious in nursing homes and in cohabiting multigenerational large families, lies in this reason.
We therefore worked out a strategy of implanting approved probiotics into the colon using our standard protocol which we used with FMT and we call this Gut Flora Replacement Therapy (GFRT). To our knowledge, this is the first time this approach has been used, but we note that Colonic Hydrotherapists regularly advertise this approach as part of their offering, but use very small amounts of implanted probiotics for a limited number of occasions. There are no published papers that can be found on this approach used by Colonic Hydrotherapists.
Every patient in this study signed an informed consent to allow us to use their information on an anonymous basis. Every patient also had a Microbiome Mapping test before and after treatment.[7-9]
This study is observational and retrospective.
Materials and Method
We start off the protocol with colonic hydrotherapy and then we do ten daily implants, five days one week, five days the next, of various bacteria outlined in Table 1.
We use a Woods Gravity Feed tank, using a closed system, and in order to administer the FMT or the GFRT we use a 40cm rectal catheter, inserted to 33 cm.
We used 16S mRNA Microbiome Mapping on all of these patients [7-9]
Table 1: List of bacterial species used in Gut Flora Replacement.
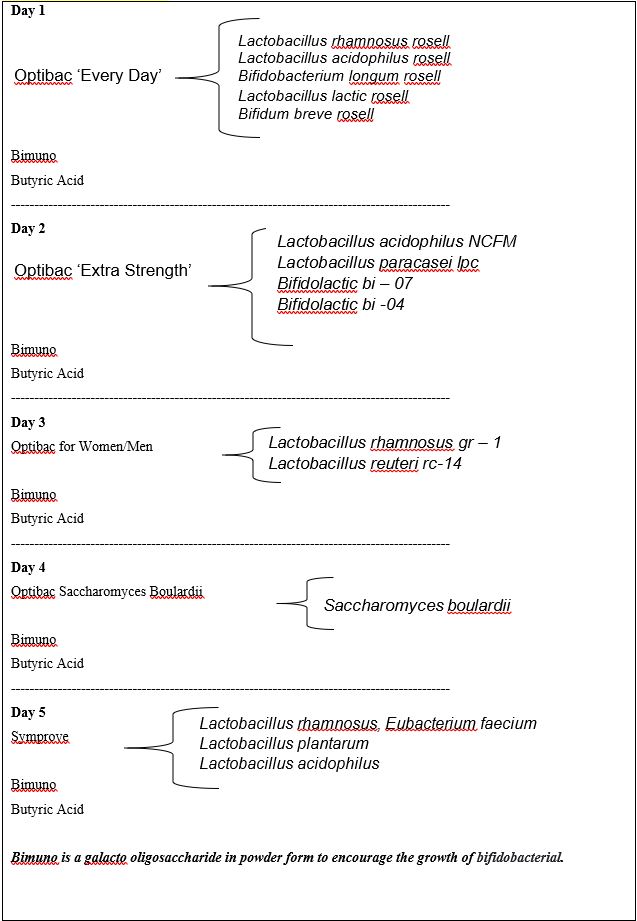
We used approved probiotic preparations as outlined in Table 2:
Table 2: Protocol for Gut Flora Replacement Therapy.

Results
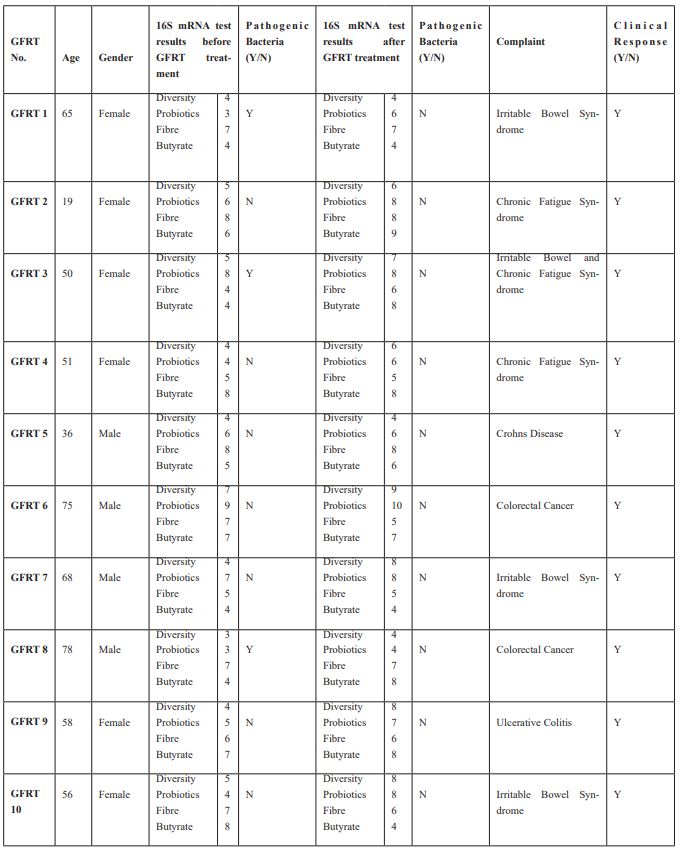
We used the data from 10 randomly chosen patients from our cohort of FMT patients and the same for 10 randomly chosen patients who had received GFRT.
We measured their microbiomes using 16S mRNA tests, the results are shown in Tables 3 and 4. The reference values for the laboratory were used for these tests are obtained from a specially selected verified subset of user’s data containing hundreds of samples which were identified as coming from healthy donors. The 16S mRNA test done on each patient before and after treatment showed no significant difference between the FMT and the GFRT groups.
Table 3: Faecal Microbiome Transplantation (FMT) Diversity, Probiotics, Fibre and Butyrate Ideal result for all of these is 8 or above.
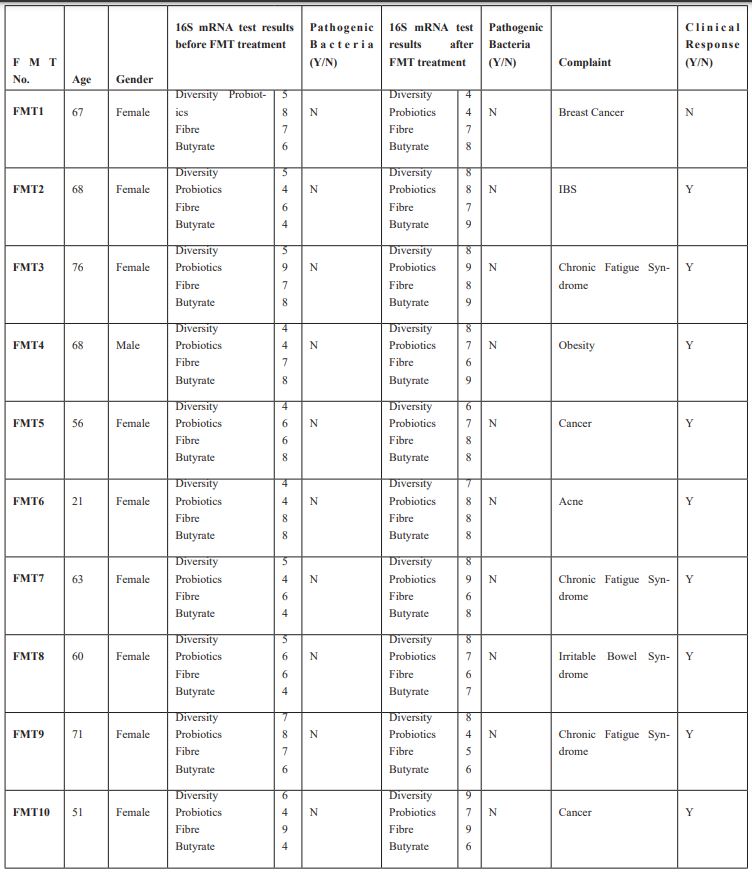
Table 4: Gut Flora Replacement Therapy (GFRT) Diversity, Probiotics, Fibre and Butyrate – Ideal result for all of these is 8 or above.
Conclusion
We have clearly demonstrated here that GFRT is a safe and effective alternative to FMT, which now carries significant risk due to the COVID-19 pandemic.
The next step forward would be a randomised control study comparing FMT to GFRT for c-difficile. The evidence for FMT being effective in c-difficile is well established [10].
The results of the 16s mRNA test before and after each treatment showed no significant difference between the FMT and GFRT groups.
References
- Shaan Gupta, Emma Allen Vercoe, Elaine O. Fecal Microbiota Transplantation: in perspective. Petrof, Therap Adv Gastroenterol 2016; 9(2): 229–239. doi: 10.1177/1756283X15607414
- Kenyon JN, Shelly Coeb, Hooshang Izadib. A retrospective outcome study of 42 patients with Chronic Fatigue Syndrome, 30 of whom had Irritable Bowel Syndrome. Half were treated with oral approaches, and half were treated with Faecal Microbiome Transplantation. Human Microbiome Journal, 2019.
- Susan Amirian E. Potential fecal transmission of SARS-CoV-2: Current evidence and implications for public health, 2020.
- Gianluca Ianiro, Benjamin H Mullish, et al. Reorganisation of faecal microbiota transplant services during the COVID-19 pandemic Correspondence to Professor Giovanni Cammarota, Digestive Diseases Center, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Università Cattolica del Sacro Cuore, Rome 00168, Italy.
- Yousra Sbaoui, Faïza Bennis, Fatima Chegdani. SARS-CoV-2 as Enteric Virus in Wastewater: Which Risk on the Environment and Human Behavior? PMID: 33795937.
- Matthew Wade, Davey Jones, Andrew Singer, Alwyn Hart, Alex Corbishley, Christopher Spence, et al. Wastewater COVID-19 Monitoring in the UK: Summary for SAGE, 2020.
- Jean Baldus Patel. 16S rRNA Gene Sequencing for Bacterial Pathogen Identification in the Clinical Laboratory
- Molecular Diagnosis 2001; 6: 313–321.
- Neethu Shah, Haixu Tang, Thomas G. Doak, Yuzhen Ye. Comparing Bacterial Communities Inferred From 16S rRNA Gene Sequencing Biocomputing 2011, pp. 165-176 (2010).
- Jethro S. Johnson, Daniel J Spakowicz, Bo-Young Hong, Lauren M Petersen, Patrick Demkowicz, Lei Chen, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis, Weinstock.
- Fecal microbiota transplantation in relapsing Clostridium difficile infection - Faith RohlkeCollege of Medicine, University of Illinois at Chicago, Chicago, IL, USA; - College of Medicine, University of Illinois at Chicago, Chicago, IL, USA and Neil Stollman - Northern California Gastroenterology Associates, 3300 Webster St, Suite 312, Oakland, CA, 94609, USA. Therap Adv Gastroenterol. 2012 Nov; 5(6): 403–420. doi: 10.1177/1756283X1245363

