Assessment of Microbes Found Near Telecommunication Mast Soil in Two Nigerian Cities
Oludare Temitope Osuntokun*, Oluyemi Joshua Oluwaseun and Ogesusi Johnson Adekunle
Department of Microbiology, Adekunle Ajasin University, Nigeria
Received Date: 05/11/2020; Published Date: 14/12/2020
*Corresponding author: Oludare Temitope Osuntokun, Department of Microbiology, Faculty of Science, Adekunle Ajasin University, Akungba Akoko, Ondo State, Nigeria. Email: osuntokun4m@yahoo.com
Abstract
Telecommunication masts are technologies that help in transmission of signal at a specific frequency. The basic objective of this research work is to assess, isolate, characterization and microbiologically identify the soil microbial flora found near telecommunication mast soil in two Nigerian cities (Oka Akoko and Owo, Ondo State). Two samples were collected from both regions (Oka and Owo) randomly and in each site, the samples were collected base on distance from the mast (0meter, 10meters, 20mters, 30mters, and 40meters). The total number of isolate was thirty four (34) for bacteria and thirty six (36) for fungi. The soil samples were serially diluted and 1ml of the dilution was transferred to separate labeled Petri dished according to Ajayi’s methodology. Pour plate techniques was adopted and the isolates were identified based on cultural physiological and biochemical characteristic cs for bacteria isolates. The fungal propagules are either hyaline (colourless) or different colours and the hyaline mycelia /spores/conidia and cytoplasm were stained using Lactophenol and cotton blue, The stained specimens were observed under the light microscope (Magnus MLXi plus) for identification and microphotographs were taken under 10X × 40X magnification for the conidia, conidiophores and arrangement of spores The physico-chemical parameters were measured by standard methods. Physical and chemical parameters of soil such as pH, salinity and organic carbon were analyzed. The bacterial isolated were Alcaligenes species, like A. paradoxus, A. xylosoxidans, and A. latus. Actinomyces bovis. Atrobacter diversus. Bacillus species such as B. pumilus, B. magaterium, B. subtili, B. cereus and B. licheniformis. Citrobacter diververseus. Cellulomovas flavigena. Corynebacterium amycolatum. Clostridium species. Such as C. tatani, C. sporogenes. Enterobacter dissolovens. Leuconostoc lactus. Listeria grayi. Micrococcus species such as M. agilis M. halobius. M. luteus. Pseudomonas species such as P. flourescens. P. alcaligenes. Streptobacillus moniformis. Sarcina ventriculi and sporolactobacillus inulinus for bacteria isolates and Fusarium equiseti, Penicillium digitatum, Aspergillus clavatus, Aspergillus niger, Rhizopus arrhizus, Wallemia sebi, Saccharomycopsis fibuliger, Fusarium avenaceum, Sporobolomyces roseus, Fusarium equiseti, Chrysosporium xerophilum, Epicoccum nigrum, Penicillium digitatum Sporobolomyces roseus for the fungi isolates. Bacillus subtilis and Aspergillus niger is the most common species found in both regions (Oka and Owo) of study. It is recommended that further study should be taken on the effect of the non-ionizing radiation of the telecommunication mast on the soil microbial flora by the adoption of sequencing methodology, to know their ecological effect and activities both on soil and plants. It is evident in this research that the microorganisms found around the telecommunication mast are normal soil flora.
Keywords: Telecommunication Mast; Bacteria; Fungi
Introduction
The purpose of this research work is to assess, isolate, characterize and identify the microbe (Bacteria and fungi) found in the near telecommunication mast soil in two Nigerian cities in Ondo state, Nigeria.
Cell phone technology has revolutionized the telecommunication sector all over the world especially in the third world country like Nigeria. Due to its several advantages, cell phone technology has grown exponentially in the last two decade. The numbers of cell phones and cell towers are increasing without giving due respect to its disadvantageous cpability (Saracci and Samet, 2010) [1]. Radiation effects are divided into two (2), thermal and non-thermal effects. Thermal effects are similar to that of cooking in the microwave oven while Non-thermal effects are not well defined but it has been reported that non-thermal effects are 3 to 4 times more harmful than thermal effects. Cell tower antennas transmit in the frequency range of 869 - 894 MHz (CDMA), 935 - 960 MHz (GSM900) and 1810 – 1880 MHz (GSM1800) (Levitt and Lai, 2010) [2]. Mobile phone operators divide a region in large number of cells, and each cell is divided into number of sectors. The base stations are normally configured to transmit different signals into each of these sectors.
In general, there may be three sectors with equal angular coverage of 120 degrees in the horizontal direction as this is a convenient way to divide a hexagonal cell (Blettner et al., 2009) [3]. If number of users is distributed unevenly in the surrounding area, then the sectors may be uneven. These base stations are normally connected to directional antennas that are mounted on the roofs of buildings or on free-standing masts (Schreier et al., 2006) [4]. The antennas may have electrical or mechanical down-tilt, so that the signals are directed towards ground level. A base station and its transmitting power are designed in such a way that mobile phone should be able to transmit and receive enough signal for proper communication up to a few kilometers (Blettn er et al., 2009) [5].
Majority of these towers are mounted near the residential and office buildings to provide good mobile phone coverage to the users. These cell towers transmit radiation 24x7, so people living within 10’s of meters from the tower will receive 10,000 to 10,000,000 times stronger signal than required for mobile communication (Blettner et al., 2009) [5].
In recent years, life has been chronically exposed to microwaves and RFR (Radiofrequency radiation) signals from various sources, including Global system for mobile communication (GSM) and UMTS/3G/4G wireless phones and base stations, Wireless Local Area Networks (WLAN), Wireless Personal Area Networks such as Bluetooth (WPAN), and DECT (Digital Enhanced (former European) Cordless Telecommunications) that are erected indiscriminately without studies of environmental impact assessments. The effects of RFR on the biological functions of living organisms which include bacteria and fungi represent an emerging area of interest with respect to environmental influences on human health.
Materials and Methods
Study sites of Near Telecommunication Mast Soil in Two Nigerian Cities.
The samples were collected from two (Oka and Owo city) different regions, Oka Akoko south west, Ondo State, Nigeria. The first site (Oka) has geographical coordinates are 70 27’ 0” North, 50 48’ 0” East. It is located 7.20 latitude and 5.59 longitude and it is situated at elevation 348 meters above sea level. The second site region under study (Owo city) situated in Ondo state, Nigeria, its geographical coordinates are 7° 11' 0" North, 5° 35' 0" East.
Method for collection of soil samples
Samples were collected near roots of telecommunication mast (up to 15cm depth), from 0m to 40m at 10m interval between each collection point where most of the microbial activity is concentrated into a sterilized polythene bags and brought to laboratory for further studies (Waksman, 2012) [6].
Isolation characterization of bacterial isolates from Near Telecommunication Mast Soil in Two Nigerian Cities
1. Serial Dilution of Soil Sample
The serial dilution method was carried out according to Ajayi et al., (2016) [7] methodology. This method, using sterile distilled water. 1g of soil sample was weighed and added to 10 ml of distilled water in conical flask to make soil suspension, the mixture was shaken to enable equal distribution of soil particles and was labelled appropriately as ‘’A’’. Before the soil settles, 1 ml of the suspension was taken with a sterile pipette and transferred to 9 ml deionized water. This was vortexed very well, and labeled as “B”. This experiment was repeated for three consecutive times, each time with 1ml of the previous suspension and 9ml deionized water. All these were labeled as tubes C, D, E. This experiment was repeated for three times, each time with 1 ml of the previous suspension and 9 ml deionized water. Label these sequentially as tubes C, D, and E. This results in serial dilutions of 10-1 through 10-5 grams of soil per ml (Ajayi etal. 2016) [7].
2. Morphological and Colony Characterization of Bacterial Isolates from Near Telecommunication Mast Soil in Two Nigerian Cities
Soil contains varieties of microorganism including bacteria and fungi that can be established in any natural environment. Bacteria are the most important and abundant microorganism which is present in surrounding environment. These are very small, unicellular, primitive and non-chlorophyll containing microorganism. Dilution method is one of most important method to isolate the soil bacterium which allows the list of living cells in the soil. An enzymatic activity of one bacterium differs from another bacterium. Biochemical test is used to differentiate among the other bacteria (Gowsalya et al., 2014). The top-three chitin degrading bacterial strains will be provisionally identified based on morphological and biochemical characteristics. The colony characteristics (margin, elevation, form, surface, color, opacity and size) on the nutrient agar, growth pattern on selective media and microscopic characteristic after staining (cell shape, Gram reaction and spore formation) will be recorded. A series of biochemical tests will be performed for identification of bacteria on basis of Bergey's Manual of Systematic Bacteriology.
3. Gram Staining of Bacterial Isolates
Gram staining is used to determine the nature of the bacterium. Colonies grown on nutrient agar will be gram stained as per the procedure explained by (Josephine et al., 2012) [8]. A slide of cell sample to be stained was made. Sample was heat -fixed to the slide by carefully passing the slide with a drop or small piece of sample on it through a Bunsen burner three times. Primary then add the primary stain (crystal violet) to the sample/slide and incubate for 1 minute. Rinse slide with a gentle stream of water for a maximum of 5 seconds to remove unbound crystal violet. Followed by the addition of Gram's iodine for 1 minute- this is a mordant, or an agent that fixes the crystal violet to the bacterial cell wall. Rinse slide with acetone or alcohol for 30 seconds and rinse with a gentle stream of water. The alcohol will decolorize the sample if it is Gram negative, removing the crystal violet. However, if the alcohol remains on the sample for too long, it may also decolorize Gram positive cells. Add the secondary stain, safranin, to the slide and incubate for 1 minute. Wash with a gentle stream of water for a maximum of 5 seconds. If the bacteria is Gram positive, it will retain the primary stain (crystal violet) and not take the secondary stain (safranin), causing it to look violet/purple under a microscope. If the bacterium is Gram negative, it will lose the primary stain and take the secondary stain, causing it to appear red when viewed under a microscope. (Josephine et al., 2012) [8].
4. Biochemical Tests for Identification of Bacterial Isolates from Near Telecommunication Mast Soil in Two Nigerian Cities
1. Preparation of Inoculum Used for Research Work
A sterile wire loop was used to pick a colon from the culture of isolated organisms and inoculated into a prepared slant and incubate. The biochemical tests was done according to method applied by (Aaisha and Barate, 2016) [9]. The staining was followed by use of various biochemical reagents and tests to get closer to the identification of bacteria. The biochemical tests that will was for the bacterial identification are carbohydrate fermentation test, enzymes test and IMViC test (Aaisha and Barate, 2016) [9].
2. Biochemical Tests
The enzymes test facilitates the detection of the enzyme in bacteria. It is essential for differentiating bacteria that can produce the particular enzyme (positive) from bacteria that are unable to produce the enzyme (negative). While it is primarily useful in differentiating between genera, it is also valuable in speciation of certain gram positives from gram-negative. The enzymes include; amylase, urease, gelatinase, catalase and oxidase. The catalase test will be carried out using the method explained by (Aaisha and Barate, 2016) [9].
Catalase Test
A microscope slide will be placed inside a petri dish. A, the petri dish cover should be available. Using a sterile inoculating loop or wooden applicator stick, small amount of organism will be collected from a well-isolated 18- to 24-hour colony and place it onto the microscope slide. Be careful not to pick up any agar. Using a dropper or Pasteur pipette, will place 1 drop of 3% H2O2 onto the organism on the microscope slide will not mix. Immediately will cover the petri dish with a lid to limit aerosols and observe for immediate bubble formation (O2 + water = bubbles). Observe for the formation of bubbles against a dark background enhances readability.
Oxidase Test
Filter paper was used for this test. A sterile loop was used to smear the isolate on the filter paper in a stained manner, while a sterile pipette was used to make a few drops of oxidase reagent (tetra-methyl-phenylenediaamine hydrochloride) (TMPD) on the spot stained with the isolate. The result shows formation of purple color within 10seconds indicating a positive reaction, while a negative result retained the color of the oxidase reagent on the filter paper (Jurtshuk et al.1976,) [10].
Imvic Test
IMViC (indole, methyl red, Voges proskauer and Citrate) procedures, a battery of tests designed to distinguish among members of the family Enterobacteriaceae.( Aaisha and Barate, 2016) [9].
Indole Test
The indole test screens for the ability of an organism to degrade the amino acid tryptophan and produce indole. It is used as part of the IMViC procedures. The indole test will be carried out using the method explained by (Aaisha and Barate, 2016) [9]. Inoculate the tube of tryptone broth with a small amount of a pure culture, Incubate at 37°C for 24 to 48 hours, To test for indole production, add 5 drops of Kovác's reagent directly to the tube,A positive indole test is indicated by the formation of a pink to red color (cherry red ring) in the reagent layer on top of the medium within seconds of adding the reagent,If a culture is indole negative, the reagent layer will remain yellow or be slightly cloudy.
Citrate Utilization
Simmons citrate medium is a nutrient substrate that offers ammonium salts as the only source of nitrogen and citrate as the only carbon source. The degeneration of citrate leads to alkalinisation of the medium which is indicated by the pH indicator bromothymol blue changing color from green to deep blue (Forbes et al., 2016) [11]. Peptone broth was prepared and inoculated with the bacterial colony and incubated for 24 hours. Simmons citrate medium was prepared by prepared by dissolving 24 grams of a Simmon Citrate.
Agar into 1liter of distilled or deionized water and homogenized. The medium was then dispensed into a clean test tubes and autoclaved at 1210C for 15minutes. They were allowed to cool and gel in a slant form. Inoculums from the incubated broth was picked and inoculated on the surface of the Agar with a sterile inoculating loop. They were then incubated for 24-48 hours. Color change from green to deep blue indicates a positive result while tubes with no color changes indicates a negative result (Forbes et al. 2016,) [11].
5. Sugar Fermentation for Bacterial Isolates from Near Telecommunication Mast Soil in Two Nigerian Cities
A situation whereby carbohydrates are utilized partially or completely in the absence of oxygen is referred to as fermentation. But sugar is utilized in the presence of oxygen such reaction is known as oxidation. Peptone water (2.0g), phenol red (0.02g), sodium chloride (2.0g) and fermentable sugar (2.0g) was weighed into a conical flask and 100ml of distilled water was added and agitated to dissolve. This preparation was decanted into different test tubes with Durham tubes inside, properly covered with cotton wool and aluminum foil with paper tape used for labeling of the tubes before sterilizing at 1210C for 15minutes. The following sugars are used: glucose, maltose, sucrose, galactose, mannitol. Inoculation of the test tube were done using fresh 18-24hours old culture of the isolates grown on nutrient agar plates and a sterile inoculate at 370C for 7days, inoculated tubes served as controls. The result was examine daily till the seventh day, changes in the indicator from red to yellow signifies positive or utilizing of sugars while production of gas was detected with inverted Durham tube (Ortiz et al.,2017) [12].
6. Carbohydrate Fermentation Test for Identification of Bacterial Isolates Near Telecommunication Mast Soil in Two Nigerian Cities
The carbohydrate fermentation test is used to determine whether or not bacteria can ferment a specific carbohydrate. Carbohydrate fermentation patterns are useful in differentiating among bacterial groups or species. It tests for the presence of acid and/or gas produced from carbohydrate fermentation. Basal medium containing a single carbohydrate source such as Glucose, Lactose, Sucrose, Mannitol, Maltose, sorbitol, fructose, xylose, ribose and inositol will be used for this purpose. A pH indicator (such as Andrade’s solution, Bromcresol purple (BCP), Bromothymol blue (BTB) or Phenol red) is also present in the medium; which will detect the lowering of the pH of the medium due to acid production. Small inverted tubes called Durham tube is also immersed in the medium to test for the production of the gas (hydrogen or carbon dioxide)..(Aaisha and Barate, 2016) [9].
Isolation characterization of Fungi isolates from Near Telecommunication Mast Soil in Two Nigerian Cities
1. Isolation of fungi from the soil samples
The isolation of fungi from soil was carried out in order to identify each fungus up to species level. The serial dilution plating method was used to dilute the soil sample as described by (Waksman, 2012) [6] with the purpose of minimizing the fungi in the soil in each dilution. From the collected soil samples, (Soil dilution method: Waksman, 2012) [6] 1g of each soil was weighed and suspended into a 10ml of sterile distilled water. 1ml of stock culture was added into a test tube containing 9ml of sterile distilled water and was diluted six times and labeled as 10-1 until 10-6. 0.5ml of the solution in the vial was pipetted into a prepared sterile petriplates containing sterile Potato Dextrose Agar the plates and were incubated for 5-7 days at 28oC (room temperature). A greater number of species were isolated most of the fungus sporulate heavily. The colonies of fungi that appeared on the plates after incubation were then isolated in a new plate. A pure culture of each colony type on each plate was obtained and maintained. The maintenance was done by sub-culturing each of the different colonies onto another new PDA plate. The colonies were incubated again at room temperature for five days (Waksman, 2012) [6].
2. Staining of fungi sample from soil sample
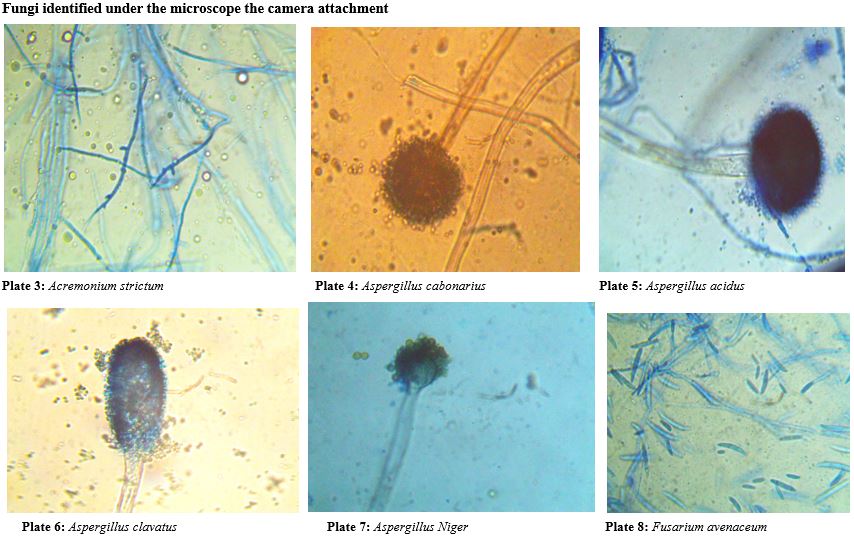
The fungal propagules are either hyaline (colourless) or different colours. The hyaline mycelia /spores/conidia and cytoplasm were stained using Lactophenol and cotton blue. Cotton blue were used to stain cytoplasm and results in light blue background. Lacto phenol acts as a cleaning agent. The stained specimens were observed under the light microscope (Magnus MLXi plus) for identification and microphotographs were taken under 10X × 40X magnification for the conidia, conidiophores and arrangement of spores (Aneja, 2018) [13]. The fungi were identified with the help of literature (Gilman, 2014.; Mc.Gill et al., 2016) [14,15].
3. Identification of fungi from soil sample
Fungal morphology were studied macroscopically by observing colony features (Colour and Texture) and microscopically.
4. Physico-chemical analysis of soil sample
The collected soil samples were characterized for its physico-chemical properties. The physico-chemical parameters were measured by standard methods. Physical and chemical parameters of soil such as pH, salinity and organic carbon were analyzed.
Moisture Contents
The analysis of the moisture contents of the samples was as described by (Dyan et al., 2013) [16]. Briefly; a porcelain crucible was placed in an oven at a temperature of 105oC and allowed to stand for 2 hours. It was allowed to cool at room temperature in desiccators. The empty crucible was weighed (empty crucible weight = x). Ten grams (10g) of sample were weighed in the crucible and weighed again (sample + crucible weight = Y). The crucible was placed for 12 hours in the oven at 105oC. The setup was allowed to cool at room temperature in a desiccator and weighed again (Z). Moisture content of the samples was calculated thus:
M (Moisture content) % = [(Y-Z) x 100%]/(Z-X)
MCF (moisture correction factor) = [100+M%]/100 (Dwell, 2012) [17].
pH Contents
The pH of the soil was measured potentiometrically in a 1:2 soil-water suspensions as described by (Dawey and Conyers, 2014) [18]. Briefly; twenty five grams (25g) air-dried, 1g sieved soil sample was weighed into a 100 ml flask, and 50 ml distilled water added and agitated for one hour. The pH meter was calibrated using pH buffer, after which the pH of the suspension was measured.
Organic Carbon Contents
The organic carbon in the sample was oxidized with potassium dichromate and sulphoric acid according to the method of (Nelson and Sommers, 2012) [19]. Briefly, one gram (1g) of the soil sample was weighed into a 500ml conical flask, ten milliliter (10ml) of 1N K2Cr2O7 and 20ml of conc.H2SO4 were added. The flask was swirled carefully and allowed to stand for 30 minutes, 200ml distilled water and 10ml H3PO4 were added slowly. One milliliter (1ml) of diphenylamine indicator was added and then titrated against 0.5N ferrous ammonium sulphate solution until green colour appeared, indicating the end point. A blank was run simultaneously and organic carbon was calculated thus:
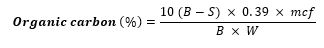
where,
B = ml of ferrous ammonium sulphate solution used for blank
S = ml of ferrous ammonium sulphate solution used for sample
mcf = Moisture correction factor
W = sample weight (g). (Waksman, 2012) [6].
0.39 = conversion factor (including a correction factor for a supposed 70% oxidation of organic carbon). According to (Schollenberger, 2013) [20], the easily oxidizable organic carbon were converted to total organic carbon by multiplying with 1.30 correction factor (or by dividing with 0.77).
Statistical analysis:
The number of colonies per plate in 1g of soil were calculated. The percentage contribution of each isolate was calculated using the formula:
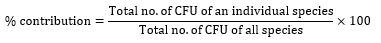
Results
Bacteria isolated soil sample found near telecommunication mast soil in two Nigerian cities. Table 1 and 2 for the bacterial isolates, a: Biochemical, Gram staining, sugar fermentation & probable identity of bacterial isolated of soil sample found near telecommunication mast soil in two Nigerian cities (Oka site 1,2 & 3).
At Oka, Ondo State, Site 1, Table 1 and 2 demonstrated bacteria isolates Identification in Oka site 1,2 and 3. This table depicts various biochemical test, Catalase test, Indole, Oxidase, Citrate and starch hydrolysis. Other features of this table include microscopy, Gram staining reaction and sugar fermentation. The sugar used were D-Glucose, Mannitol, Maltose, Galactose and Sucrose. it was observed that all isolates shows positive reaction to sugar hydrolysis between 0 to 40 m, catalase positives at 0,10,30 and 40meter and catalase negative at 20 meter, indole negative between 0 and 40 meter,citrate positive at 0, 10, and 40metre,. Microscopically, rod shaped isolates at 0,20 and 40 meter and cocci at 10 and 20metre respectively. The Gram staining reaction at 0 meter, isolates were Gram negative rods, Gram +ve cocci at 10 and 20 meter, Gram +ve rod isolates at 30 meter and Gram +ve rod at 40meter. It was also observed that all isolates react to sugar fermentation positively and this were the probable identity of isolates, Citrobacter diverseus, Micrococcus leteus, Sarcina ventriculi, Bacillus subtilis, Pseudomonas fluorescens
At Oka site, Ondo State, 2, all isolates shows positive reaction to sugar hydrolysis between 0 to 40 m, Citrates positive at 0,20,30 and 40 meter, negative at 10m, Indole negative between 0 and 40 meter, Oxidase positive at 10m and negative at 0,20 and 30 meter, Citrate positive at 0 to 40m, Cocci isolates at 20 meter, rod at 0,10,30 meter and 40meter, Gram positive at 0,10,20 and 40 meter, negative at 20meter.it was observed that Gram + ve rod at 0 ,10,30 and 40 meter, but Gram –ve cocci at 20 m. It was also observed that all isolates react to sugar fermentation positively and this were the probable identity of isolates were Bacillus subtilis, Clostridium sporogenes, Alcaligenes paradoxus, Lystaria grayi and Bacillus magatarium
At Oka site 3, It was also observed that all isolates react to sugar fermentation positively and this were the probable identity of isolates, Pseudomonas alcaligenes, Leuconostoc lactis, Bacillus licheniformis, Clostridium tatani and Citrobacter diversus
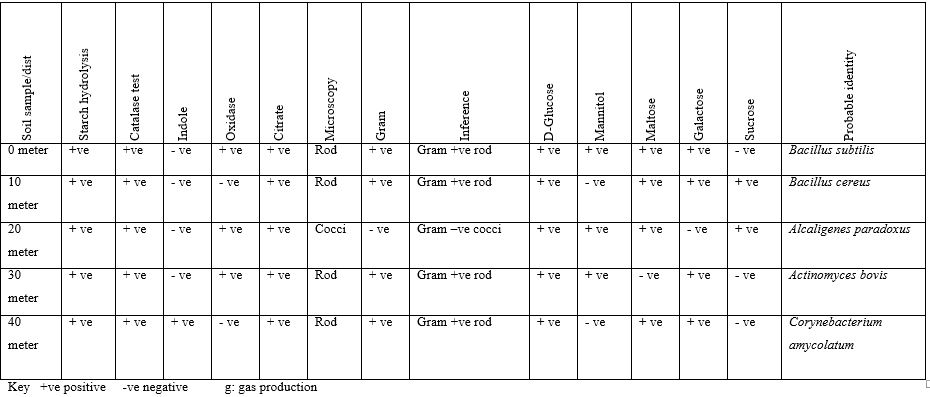
Table 3 and Table 4: Biochemical, Gram Staining and probable identity bacterial isolated of soil sample found near telecommunication mast soil in two Nigerian cities (Owo Site 1,2&3).
At Owo, Ondo State, site 1, all isolates shows positive reaction to sugar hydrolysis between 0 to 40 meter, Citrates positive at 0,10, 20 and 40meter, negative at 30 meter,, Indole negative between 0 and 40 meter, oxidase positive at 0 and 10 meter, and oxidase negative at 20,30 and 40 meter,. Citrate positive at 0,10 and 20 negative at 30 and 40 meter,, cocci under the microscopic at 0,20 and 40 meter, and rods at 20 and 30 meter. Gram positive at 0, 10 and 20 meter,,negative at 30 and 40 meter,, . It was also observed that all isolates react to sugar fermentation positively and this were the probable identity of isolates, Micrococcus halobis, Bacillus pumilus, Sporolactobacillus inulinus, Streptobacillus moniliformis and Alcaigenes xylosoxidans.
At Owo, Ondo State, site 2, all isolates shows positive reaction to sugar hydrolysis between 0 to 40 meter,, Citrates positive at 0,20,30 and 40 meter,, negative at 10 meter,. oxidase positive at 0, negative at 10, 20 and 30 meter,, , Indole negative between 0 and 40 meter,, Citrate positive at 0, 20, 30 and 40 meter,, negative at 10 meter,, Cocci under the microscopic at 10 meter and rods at 0,20,30 and 40 meter,. They were Gram positive at 0,10 and 20 meter,,negative at 30 and 40 meter,, positively and this were the probable identity of isolates, Pseudomonas alcaligenes, Leuconostoc lactis, Bacillus licheniformis, Clostridium tatan, Citrobacter diversus.
At Owo,Ondo State, site 3, It was also observed that all isolates react to sugar fermentation positively and this were the probable identity of isolates, Bacillus subtilis, Bacillus cereus, Alcaligenes paradoxus, Actinomyces bovis and Corynebacterium amycolatum.
Fungi isolated soil sample found near telecommunication mast soil in two Nigerian cities. The fungi isolates were depicted in table 5 and 6 demonstrated with Owo site 1,2 and 3. According to the findings from this study, it shows that fungal diversity of soil depends on large number of factors of the soil such as pH, organic content, and moisture. Physico-chemical analysis of soil showed that pH range of soil conditions ranging from 5.1 to 7.5 and soil textures determined the fungal population and their diversity in various telecommunication mast site. During the investigation period, 40 fungal colonies of 15 fungal species were observed. Among the isolates the genera Aspergillus and Fusarium were dominant (Table 5 & 6).
Table 5 and Table 6: Physico-chemical properties of soil samples isolated soil sample found near telecommunication mast soil in two Nigerian cities (Owo Site 1,2 &3). Table 5 and 6 depicts several parameter for the identification of fungi isolates. The parameters are Soil colour, Soil type, pH, Salinity and Organic carbon. It was observed the colour of the soil differs which include Grey, Brown and Light Grey from distance 0 to 40metre, pH ranges between 5.1 to 7,5, Salinity ranges between 0.21 to 0,37 for Owo site 1 and 3. Owo site 3 depicts soil colour between brown to Dark Grey,pH were between 5.4 to 7.2, salinity were between 0.32 to 0.70.
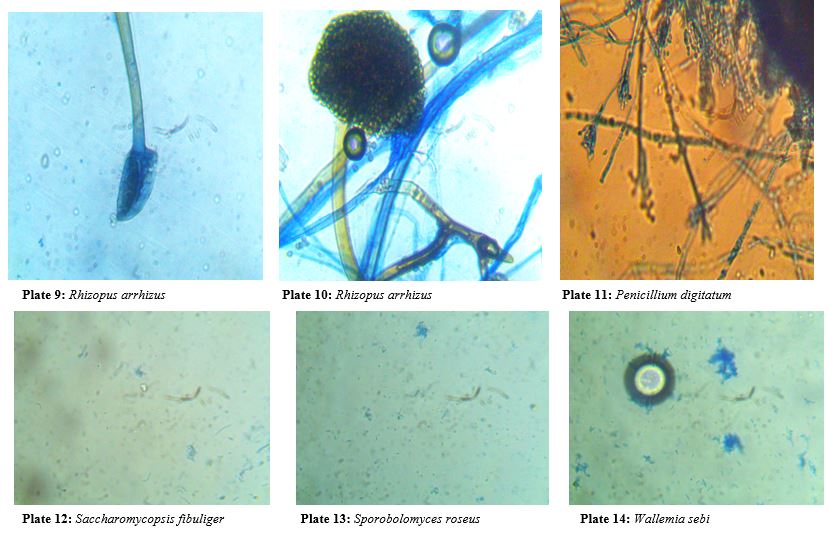
Table7. Probable fungi identity of soil samples isolated soil sample found near telecommunication mast soil in two Nigerian cities (Oka &Owo). Colour, Texture, Growth rate, Shape, Hyphae were used as the mode of identification for the two site Owo and Oka. This were the probable identity of isolates based on the parameter of identification. Aspergillus acidus, Aspergillus clavatus, Aspergillus fumigates, Aspergillus niger, Botrytis cineria, Byssochlamys nivea, Chrysosporium xerophilum,, Epicoccum nigrum,, Fusarium avenaceum, Fusarium equiseti, Penicillium digitatum, Aspergillus clavatus, Aspergillus niger, Rhizopus arrhizus, Wallemia sebi, Saccharomycopsis fibuliger, Fusarium avenaceum, Sporobolomyces roseus and Fusarium equiseti.
Table 8. Percentage frequency distribution of fungi of soil samples Isolated Soil Sample found Near Telecommunication Mast Soil in Two Nigerian Cities (Oka & Owo). Aspergillus unguis has the highest percentage (17.3%), followed by Aspergillus acidus (13.2%), Aspergillus fumigatus (11.2%), Fusarium avenaceum (7.1%), Botrytis cineria has the lowest percentage (3.0%), Byssochlamys nivea (3.0%) respectively.
Bacteria Isolated Soil Sample found Near Telecommunication Mast Soil in Two Nigerian Cities
Table 1: Biochemical, Gram Staining, Sugar Fermentation and Probable Identity of Bacterial Isolated of Soil Sample Found Near Telecommunication Mast Soil In Two Nigerian Cities (Oka Site 1 & 2).
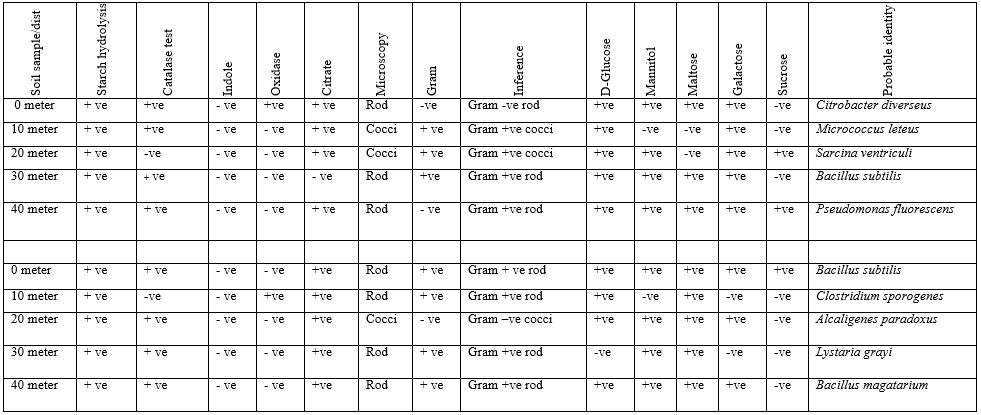
Table 2: Biochemical, Gram Staining, Sugar Fermentation and Probable Identity of Bacterial Isolated of Soil Sample Found near Telecommunication Mast Soil in Two Nigerian Cities (Oka site 3)
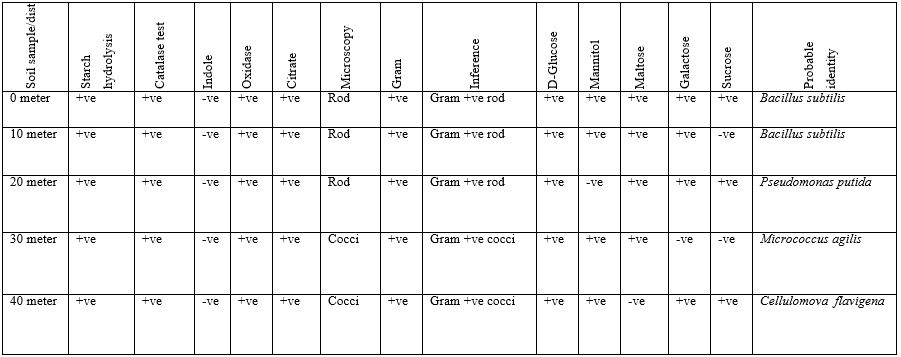
Table 3: Biochemical, Gram Staining and Probable Identity Bacterial Isolated of Soil Sample Found Near Telecommunication Mast Soil In Two Nigerian Cities (Owo Site 1 & 2).
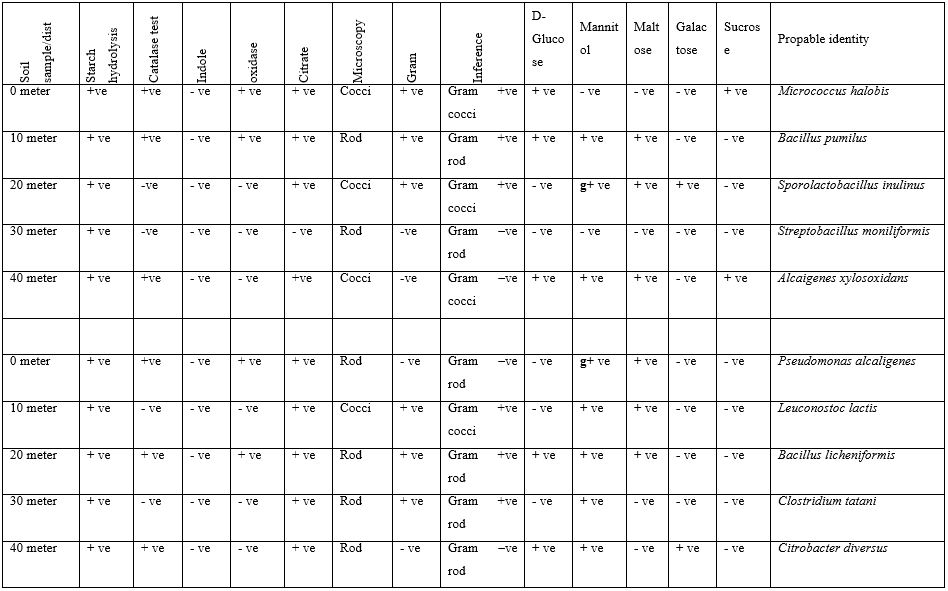
Table 4: Biochemical, Gram Staining and Probable identity Bacterial Isolated Soil Sample found Near Telecommunication Mast Soil in Two Nigerian Cities (Owo site 3).
Fungi Isolated Soil Sample found Near Telecommunication Mast Soil in Two Nigerian Cities
Table 5: Physico-chemical properties of soil samples Isolated Soil Sample found Near Telecommunication Mast Soil in Two Nigerian Cities (Owo Site 1 &2).
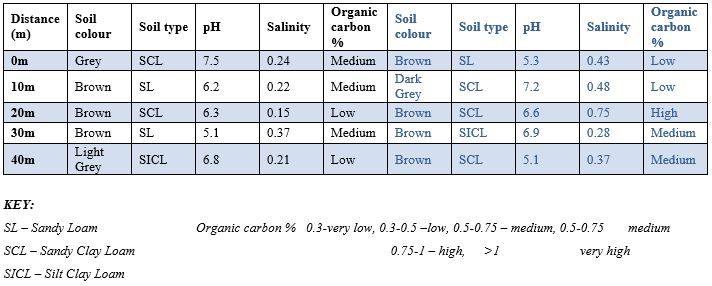
Table 6: Physico-Chemical Properties of Soil Samples Isolated Soil Sample Found Near Telecommunication Mast Soil In Two Nigerian Cities (Oka site 3).
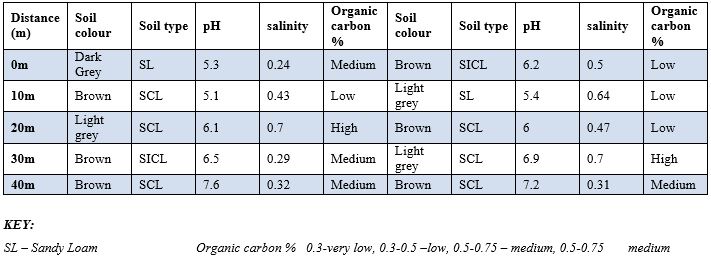
SCL – Sandy Clay Loam 0.75-1 – high, >1 very high
SICL – Silt Clay Loam
Table 7: Probable Fungi Identity of Soil Samples Isolated Soil Sample Found Near Telecommunication Mast Soil In Two Nigerian Cities (Oka &Owo).
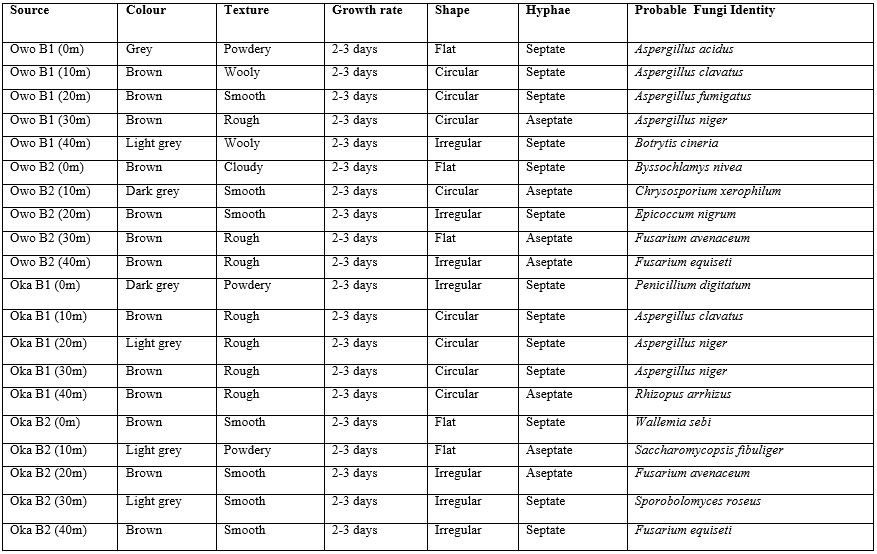
Table 8: Percentage Frequency Distribution of Fungi of Soil Samples Isolated Soil Sample Found Near Telecommunication Mast Soil In Two Nigerian Cities (Oka & Owo)
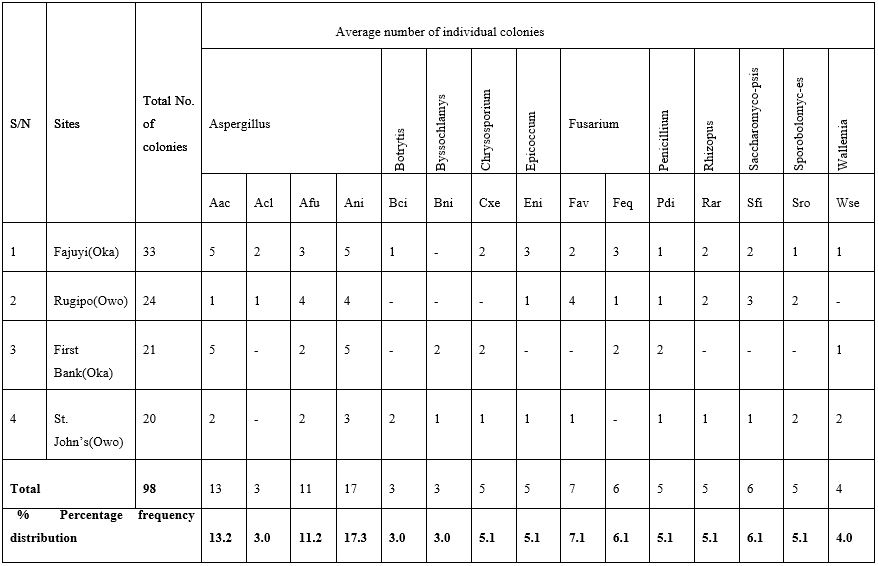
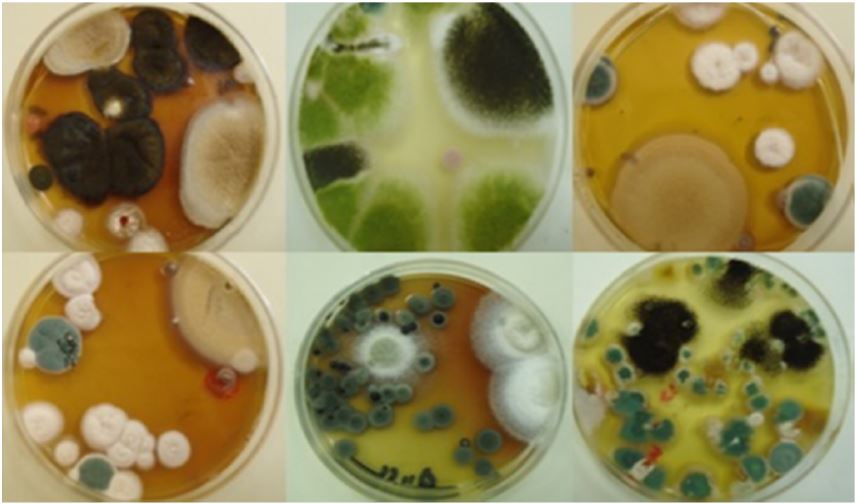
Plate 2: A number of fungal colonies isolated from the soil samples of the studied sites.
Discussion
The basic objective of this research work is to assess, isolate, characterization and microbiologically identify the soil microbial flora found near telecommunication mast soil in two Nigerian cities (Oka Akoko and Owo, Ondo State).
Microorganisms are ubiquitous and are more abundant in the soil. The following microorganisms where identified and microbiologically characterized by the use of colony morphology, biochemical reaction and Gram staining procedure during the course of this research. They are Alcaligenes species, like A. paradoxus, A. xylosoxidans, and A. latus. Actinomyces bovis. Atrobacter diversus. Bacillus species such as B. pumilus, B. magaterium, B. subtili, B. cereus and B. licheniformis. Citrobacter diververseus. Cellulomovas flavigena. Corynebacterium amycolatum. Clostridium species. Such as C. tatani, C. sporogenes. Enterobacter dissolovens. Leuconostoc lactus. Listeria grayi. Micrococcus species such as M. agilis M. halobius. M. luteus. Pseudomonas species such as P. flourescens. P. alcaligenes. Streptobacillus moniformis. Sarcina ventriculi. And sporolactobacillus inulinus.
Most bacteria isolated are cosmopolitan of which Bacillus species took the higher percentage (Nickerson et al., 2000,) [21]. In the attempt to adapt to the radiation effect in their environment, the DNA of the microorganisms become altered and mutated. (Nickerson et al., 2000.) [21]. Non-Ionizing Radiation (NIR) refers to radiative energy that, instead of producing charged ions when passing through matter, has sufficient energy only for excitation. Nevertheless it is known to cause biological effect. Some of the source of Non-ionizing radiation include telecommunication mast, telephone, EMF, power line, microwave, broadcasting antenna. The microorganisms isolated during the course of this research are the same as those isolated from ionizing radiation environment. Microorganisms are very sensitive when they are ionic radiated, Slow bacteria are very resistant when they are exposed to ionic radiation, gram negative bacteria are very sensitive, so do yeasts and molds. Micrococcus radiodurans are very resistant, and Clostridium botulium is also resistant to radiation and differs to that of Cl. sporogens, which is resistant only when it is effected by thermal treatment.
One of the most important properties of irradiation is inactivation of microorganisms, especially pathogens. Those microorganisms which are efficient in repairing their DNA show the greatest probability of survival, and are therefore the most resistant to irradiation. Since the microorganisms found in ionizing environment are the same as those isolated in this research. There is high probability that the radiation from both ionizing and non-ionizing pose No significant effect on the microorganisms. Only that the range of this effect will differ from each other due to differences in frequency and wavelength.
The effects of radiofrequency radiation on the biological functions of living organisms represent an emerging area of interest with respect to environmental influences on human health. In latest years, several studies have been performed to verify direct effects exerted by such radiations on cell functions. Although results have been somewhat controversial and not conviction, a variety of cell responses have been observed involving proliferation and differentiation (Huang et al., 2006; Lisi et al., 2006) [22, 23] gene expression (Goodman et al., 2009) [24], modulation of the membrane receptors functionality (De-Mattei et al., 2009) [25], apoptosis, alteration in ion homeostasis (Iorio et al., 2011) [26], and free radicals generation (Simk, 2007) [27].
In particular, it has been demonstrated that radiation can negatively (Justo et al., 2006) [28] or positively (Cellini et al., 2008; Belyaev, 2011) [29,30] affect functional parameters (cell growth and viability) and bacteria antibiotic sensitivity depending on physical parameters of the electromagnetic field (frequency and magnetic flux density), (Bush et al., 2011) [31].
The fungi identified during the course of this research are: Aspergillus species, Penicillium species, Chrysosporium species, Epicoccum species, Fusarium species, Wallemia species, Byssochlamys species, Acremonium species, Rhizopus species, Saccharomycopsis species, Sporobolomyces species. All these were identified from the soil samples. The result presented in this project work shows clearly the presence of fungi in soil around telecommunication masts. Results recorded shows virtually that the soil samples are acidic and is rich with both macro and micro nutrients which is favorable for the growth of fungi. Fungal diversity of any soil depends on a large number of factors of the soil such as pH, organic content and moisture (Rangaswami et al., 2008) [32].
Total number of colonies, CFU/gm of soil, Percentage of frequency of fungi. The colony forming units of fungi varied from minimum of 1.56 to maximum of 18.75x 10-2/-3CFU/gm. The percent contribution of the individual species to the total fungal population showed variation. The maximum percent contribution showed by Aspergillus unguis with 17.3%, followed by Aspergillus acidus with 13.2%, Aspergillus fumigatus 11.2%, Fusarium avenaceum 7.1%, to the least, Botrytis cineria 3.0%, Byssochlamys nivea 3.0% in order of dominance.The results from the fungi identification is closely related with report of soil fungi isolation by Zaman et al., (2012) [33]. Physicochemical parameters of soil such as pH, moisture content, macro nutrients (organic carbon, salinity) were observed in this research work, it was observed that the soil near the telecommunication mast contains what it needed for all forms of life like microbes to survive, in regardless of the radiation from the telecommunication mast.
In conclusion, it should the reiterated that the amount of radiation from the mast is not enough to destroy the nearby microbes. But with further research, it may have affected their genetic code if this is applicable, this is left for future research analysis where the composition of genetic code of isolated microbe is study to know if thete is changes in their genetic make-up either destroy, delete nor disorientated.
Competing Interests
Authors have declared that no competing interests exist.
Acknowledgement
The laboratory staff of Department of Microbiology, Faculty of Science, Adekunle Ajasin University, Akungba Akoko, Ondo State, Nigeria and Federal Medical Center laboratory section, Owo, Ondo state, Nigeria.
References
- Saracci R, Samet J. Commentary: Call me on my mobile phoneor better not?—a look at the INTERPHONE study - International Journal of Epidemiology, published online on 2010; 39(3):695-698.
- Levitt B, Lai H. Biological effects from exposure to electromagnetic radiation emitted by cell tower base stations and other antenna arrays, Rev. 2010; 18: 369–395.
- Blettner M, Schlehofer B, Breckenkamp J, Kowall Schmiedel S. Mobile phone base stations and adverse health effects: phase 1 of a population-based, cross-sectional study in Germany. Occup Environ Med; 2009; 66: pp 118–23.
- Schreier N, Huss A, Röösli M. The prevalence of symptoms attributed to electromagnetic field exposure: a cross-sectional representative survey in Switzerland. Soz Praventivmed; 2006; 51: pp 202–209.
- Blettner M, Schlehofer B, Breckenkamp J, Kowall Schmiedel S. Mobile phone base stations and adverse health effects: phase 1 of a population-based, cross-sectional study in Germany. Occup Environ Med; 2009; 1 66: pp 118–23.
- Waksman SA. Principles of Soil Microbiology. Williams and Wilkins Co. Baltimore, Md. 2012; pp. 1-654.
- Ajayi EIO, Ogungbuji ET, Ganiyu N. Analgesic potential of certain traditional African herbal extracts in high fat dietmanipulated hyperglycaemic rats. Ife Journal of Science 2015; 17(2): 511 – 517.
- Josephine SF, Ramya VS, Neelam D, Suresh BG, Siddalingeshwara KG, Venugopal N, et al. Isolation, production and characterization of protease from Bacillus Sp isolated from soil sample Microbiol. Biotech. Res. 2012; 2 (1):163-168
- Aaisha GA, DL Barate. J.Curr.microbial.App.Sci, 2016; 5(1):514-524.
- Jurtshuk P, McQuitty DN. Use of oxidase test for characterizaing oxidative metabolism in bacterial. Applied Environmental Microbiology. 1976; 31: 688-679.
- Forbes AB, Daneil FS, Alice SW. Bailey and scotts. Diagnosis Microbiology (10 ed.) 2016; Pp. 430.
- Ortiz ME, Bleckwedel J, Fadda S, Picariello G, Hebert EM. Global analysis mannitol 2-Dehydrogenase Lactobacillus reuteri CRL 1101 during Mannitol production through enzymatic, Genetic and proteomic approaches PLos ONE 2017; 12(1).
- Aneja DL. Fungal diversity and its implications for genetic resource collections. Studies in Mycology. 2018; pp: 5-19
- Gilman V. Seasonal Distribution of soil fungi and chemical properties of montane wet temperate forest types of Tamilnadu. African Journal of Plant science. 2014; 4(6):190-196.
- Gill TB, Prasad BK, Mishra N. Influence of soil environment and surface vegetation on soil micro flora in a coastal sandy belt of Orissa. India Journal of Human Ecology. 2016; 27(1):69-73.
- Dyan DM, Hartel PG, Fuhrmann JJ, Zuberer DA. Principles and Applications of Soil Microbiology. (2nd edition). Edited by David M. Sylva, Pearson Prentice Hall, Upper Saddle River: New Jersey. 2013.
- Dwell CW. Isolation, culture and maintenance of endophytic fungi of grasses In: Isolation of biotechnological organisms from nature. Edition by Labeda, P., McGraw- Hill, New York, USA. 2012.
- Dawey GF, Conyers JD. Abundance and diversity of microfungi in leaf litter of a lowland rain forest in Costa Rica. Mycology, 2014; 86:187-198.
- Nelson K, Sommers, N. Seasonal distribution and abundance of soil fungi from forest soils of wet evergreen forest of Tamil Nadu, Journal of modern biotechnology. 2012; 1(1):8-14.
- Schollenberger SB. Ecological factors governig the distribution of soil micro fungi in some forest soils of Saugar. Journal of Indian Botany society. 2013; 34:262-298.
- Nickerson CA, CM Ott, SJ Mister, BJ Morrow, L Burns-Keliher DL Pierson. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2000; 68:3147–3152.
- Huang L, Dong L, Chen Y, Qi H, Xiao D. Effects of sinusoidal magnetic field observed on cell proliferation, ion concentration, and osmolarity in two human cancer cell lines. Electromagnetic Biology and Medicine. 2006; 25: 113– 126.
- Lisi A, Ledda M, Rosola E. extremely low frequency electromagnetic field exposure promotes differentiation of pituitary corticotrope-derived AtT20 D16V cells. Bioelectromagnetics. 2006; 27: 641–651.
- Goodman R, Lin-Ye A, Geddis MS. Extremely low frequency electromagnetic fields activate the ERK cascade, increase hsp70 protein levels and promote regeneration in Planaria. International Journal of Radiation Biology. 2009; 85: 851–859.
- De-mattei M, Varani K, Masieri FF. Adenosine analogs and electromagnetic fields inhibit prostaglandin E2 release in bovine synovial fibroblasts. Osteoarthritis and Bioelectromagnetics. 2009; 29: 302–311.
- Iorio R, Delle-Monache S, Bennato F. Involvement of mitochondrial activity in mediating ELF-EMF stimulatory effect on human sperm motility. 2011; 32: 15–27.
- Simk´M. Cell type specific redox status is responsible for diverse electromagnetic field effects. Current Medicinal Chemistry. 2007; 14:1141–1152.
- Justo O R, Perez V H, Alvarez DC, Alegre RM. Growth of Escherichia coli under extremely low-frequency electromagnetic fields. Applied Biochemistry and Biotechnology. 2006; 134: 55–163.
- Cellini L, Grande R, Di-Campli E. Bacterial response to the exposure of 50Hz electromagnetic fields. Change Biology 2008; 18: 1918–1927.
- Belyaev I. Toxicity and SOS-response to ELF magnetic fields and nalidixic acid in coli cells. Mutation Research. 2011; 722: 56–61.
- Bush K, Courvalin P, Dantas G. Tackling antibiotic resistance. Nature Reviews Microbiology. Vol. 9, pp. 894–896, 2011; 17: 252–262.
- Rangaswami VN, Prakash P, Manoharachary C. Fungistasis in forest soil using the dominant soil fungi. Biome. 2008; 2(1):126- 128.
- Zamar CB, Jin ZX, Li JM. Diversity of bacterial physiological groups and microbial flora in the soil of eight forest types of Tiantai Mountain, Zhejiang. Biodiversity Science. 2012; 9(4):382-388.

