Ocimum Gratissimum Capped Sulfur Nanoparticles and Their Antibacterial Efficacy Against Multidrug-Resistant Microbes
Bankole OM, Osuntokun OT*, Adeola A, Owoeye A
1Centre for Bio-Computing and Drug Development (CBDD), Adekunle Ajasin University, Nigeria
2Department of Microbiology, Adekunle Ajasin University, Nigeria
3Department of Chemical Sciences, Adekunle Ajasin University, Nigeria
Received Date: 09/09/2020; Published Date: 11/11/2020
*Corresponding author: Oludare Temitope Osuntokun Department of Microbiology, Faculty of Science, Adekunle Ajasin University, Akungba Akoko, P.M.B 001, Ondo State, Nigeria. Tel: Tel: +234 806 381 3635; E-mail: osuntokun4m@yahoo.com
Abstract
This manuscript reports for the first time synthesis of sulfur nanoparticles prepared from thiosulphate pentahydrate () using either oxalic acid alone (SNP-1), or mixture of oxalic acid and aqueous solution of Ocimum gratissimum (SNP-2). The synthesized sulfur nanoparticles were obtained in satisfactory yields, and characterized with techniques such as UV-Vis, XRD, SEM, EDX, TEM, and FT-IR. Ptrresence of capping agents: oxalic acid and biomolecule contents of Ocimum gratissimum were confirmed by FTIR. Crystallinity, morphology, shapes and elemental compositions of as-prepared nanoparticles were confirmed by XRD, SEM, TEM and EDX, respectively. Antimicrobial activities of the prepared sulfur nanoparticles against five (5) multidrug-resistant microbes were used for this research work this include Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa and Salmonella pullorum. The zone of inhibition of the sulfur nanoparticles tested against selected clinical isolates. Staphylococcus aureus was observed to have the highest susceptibility to Sulfur Nanoparticles (SNP1) mediated with Ocimum gratissimum plant extract with diameter of 20.00 mm; Escherichia coli and Salmonella eserach Pseudomonas aeruginosa a showed resistance. All tested clinical isolates were resistant to the other Sulfur Nanoparticles (SNP2) synthesized in the absence of Ocimum gratissimum plant extract.
Key words: Sulfur nanoparticles; Ocimum gratissimum; Antibiotics; Antimicrobial activity; Multidrug-resistant microbes
Introduction
Nanothechnology which spans across all areas of science and engineering deals with the fabrication, development, characterization and application of small-sized devices or materials whose sizes ranging from 1 - 100 nm [1,2]. The small-sized materials otherwise known as nanoparticles have intriguing features which differ biologically, chemically and physically from the bulk materials [2]. Due to their small sizes and large active surface areas, nanomaterials have been found useful as potent antimicrobial therapies and in other biological applications [2]. Manipulation of nanomaterials for enhanced treatment of pathogenic bacteria and other infectious diseases has continued to revolutionize global human health [2]. Metallic nanoparticles have been in the forefront and extensively reported antibacterial agents for treating bacterial infections and other multi-resistant pathogens [3,4]. Although much successes have been recorded with the use of metal-based nanoparticles as antibiotics, however they are not only harmful to the targets bacterial but are also toxic to normal human cells [5]. Thus, research attention has been shifted to the use of metal-free or non-metallic nanoparticles as therapies against bacterial infections. Sulfur nanoparticles are non-metallic nanoparticles and have been reported to have wide potential applications in agriculture, biomedical and medicine [6-8]. Rhimet al. [9], demonstrated that chitosan-capped sulfur nanoparticles exhibited strong antimicrobial activities against Gram-positive and Gram-negative bacteria; and also prevented proliferation of cancer cells with minimal toxic effect on normal cells. In another report, enhanced bactericidal and bacterial static effects of biosynthesized sulfur nanoparticles using extract of different medicinal plants against Gram-positive and Gram-negative bacteria have also been reported [10]. This is an indication that antibacterial activity of sulfur nanoparticles were enhanced in the presence of active biomolecules used as capping agents. Other biologically synthesized sulfur nanoparticles with significant bioactivities have been reported in the literature [7,8,11]. Scientists and biomedical experts are investing in biosynthetic method of preparation of nanoparticles using leaf extract of medicinal plants. Biosynthetic method have advantages over conventional chemical methods because it is eco-friendly, cost-effective, require lesser reaction time, and they do not produce hazardous by-products. Herein, biosynthetic method was employed to prepare sulfur nanoparticles using leaf extract of Ocimmum gratissimum as reducing and capping agent. The Ocimum gratissimum, also known as scent leaf or African Basil, is a perennial herbaceous plant belonging to the family of Lamiaceae, and native to Africa and Southern Asia [12]. Aqueous and alcohol extract of leaves of Ocimum gratissimum have medicinal values which has been reported to possess significant antibacterial and antifungal activities against different pathogens [12,13]. The use of leaf extract of Ocimum gratissimum for synthesis of sulfur nanoparticles have not been reported before in the literature. In this present work, sulfur nanoparticles are prepared using either only oxalic acid (abbreviated as SNP-1), or mixture of leaf extract of Ocimum gratissimum and oxalic acid (abbreviated as SNP-2), as reducing and capping agents. Antimicrobial activities of prepared nanoparticles against pathogenic microorganisms; Gram positive and Gram negative bacterial are carried out and the results are compared.
Materials and Methods
Materials
Sodium thiosulfate pentahydrate (99%), citric acid ( AR) were purchased from Sigma–Aldrich. Absolute ethanol was purchased from Pascal Scientific Limited, Nigeria, and used as received. Fresh leaves of Ocimum gratissimum were collected from campus of Adekunle Ajasin University, Akungba and used as a source of nanoparticles preparation. Deionized water was used throughout the experiments.
Method
Preparation of leaf extract: The fresh leaves of Ocimum gratissimum were removed from their stalks and washed thoroughly with running tap water to remove any attached particles or debris and finally washed with deionized water. Subsequently, the leaves were allowed to dry for 3-4 weeks at room temperature to remove the adsorbed moistures. The dried leaves were ground into fine powders in a clean agate mortal and stored in a tight container for further use. Biomolecule contents of the leave were extracted by adding 20 g of the powdered leaves to 100 mL deionized water in a 250 mL beaker, and heated to 100 °C for 60 min. The crude greenish extract was filtered through Whatmann No. 1 filter paper and stored in the refrigerator at -10 °C prior to the preparation of sulfur nanoparticles.
Biosynthesis of Sulfur Nanoparticles (SNP-1 and SNP-2), Scheme 1: Synthesis of sulfur nanoparticles in the presence or absence of Ocimum gratissimum plant extract is as follows: sodium thiosulfate pentahydrate (0.403 M) and 50 mL of Ocimum gratissimum leaf extract were to 150 mL deionized water and allowed to stir on a magnetic stirrer at room temperature for 30 min. Then aqueous solution of citric acid (2.42 M, 50 mL) was added drop wise under stirring to allow the precipitation of sodium thiosulfate as sulfur nanoparticles and SO2 Scheme 1, in accordance with previous report [14]. The mixture was stirred for additional 1 hrs and was allowed to stand undisturbed for 5 hrs for complete disproportionation of sodium thiosulfate. Biosynthesized sulfur nanoparticles was collected by centrifugation, washed with deionized water and Et OH, and dried in an oven at 50 °C for 24 hrs. For comparison, sulfur nanoparticles in the absence of leaf extract was also prepared using Na2S2O3 and citric acid but without leaf extract. Sulfur nanoparticles prepared in the presence and absence of plant extract are named SNP-1and SNP-2, respectively.
Na2S2O3(aq)+H+(aq, citricacid)?SO2(g)+S?+H2O(1) (1)
Material Characterizations
The solid reflectance spectra of prepared SNP-1 and SNP-2 were recorded on a Shimadzu UV-VIS-NIR Spectrophotometer UV-3100 with a MPCF-3100 sample compartment with samples mounted between two quartz discs which fit into a sample holder coated with barium sulfate. The spectra were recorded over the wavelength range of 800-250 nm, and the scans were conducted at a medium speed using a 20 nm slit width. Infra-red spectra were recorded on a Thermo Fisher Scientific FTIR spectrophotometer, using pressed KBr pellets. Transmission Electron Microscope (TEM) image was recorded using a JEOL JEM-2010 electron microscope operating at 200 KV. Surface morphology and elemental composition of sulfur nanoparticles were analysed using Scanning Electron Microscope (SEM) equipped with energy dispersive analysis of X-ray equipment (EDAX) (XL 30 FEG ESEM).
Sample Collection Of Clinical Isolates
Pure cultures of Five (5) selected clinical isolates; Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa and Salmonella pullorum were collected from the Microbiology laboratory of Adekunle Ajasin University for this study. These isolates were pathogenic and have been known to cause human and poultry diseases. All the culture were grown and maintained in nutrient broth at 37 °C and used for further studies.
Antibiotic Susceptibility Test For The Bacterial Isolates
The test was performed to determine the phenotypic resistance of the bacterial isolates to commonly used antibiotics. These tests were carried out following the Kirby-Bauer disc diffusion method of (CLSI, 2009). Inoculum from culture of bacteria isolates on nutrient agar slants were inoculated into test tubes containing sterilized nutrient broth and incubated at 37 °C for 18 hrs which serve as the stock for the test. Mueller-Hinton agar was prepared and sterilized, then dispensed into sterilized Petri dishes. The plates were allowed to cool for about 15min so as to allow it to gel and excess surface moisture to be absorbed.
The inoculum was introduced into plates by streaking before applying the antibiotics impregnated discs. Two types of discs were used; Cephalosporin antibiotic discs (Oxoid); Cefuroxime (30 µg), Ceftazidime (30 µg), Cefoxitin (30 µg), Cefpodoxime (10 µg), Cefepime (30 µg) and Multi-test Predetermined commercial Gram negative and Gram positive discs which were applied to the surface of the well labeled inoculated agar plated aseptically using sterile forceps. The discs were then placed firmly by slightly pressing on the inoculated plates with the sterilized forceps to ensure complete contact with the agar. After 24 hrs of incubation, each plates was examined, susceptibility to each antibiotics were indicated by a clear zone. The zone of inhibition were measured using a calibrated ruler was held on the back of the inverted petri plate and was recorded (Osuntokun et al., 2019).
Precaution taken includes:
Standardized inoculum was streaked on the plates.
Aseptical introduction of disc avoiding dragging on agar surface.
Avoidance of undiluted over-night cultures
Result and Discussions
Material Characterizations
The solid reflectance spectra of as-prepared of SNP-1 and SNP-2are shown in Figure 1. Formation of as-prepared bare and Ocimum gratissimum stabilized sulfur nanoparticles were confirmed by the presence of primary and secondary absorption peaks around 269-272 nm, and 310-312 nm, respectively. The primary absorption peaks (SNP-1; 272.5 nm, and SNP-2; 269 nm, Table 1) are due to successful disproportionation of sodium thiosulfate to sulfur nanoparticles in the presence of organic acid [15,16]; while the secondary peaks (SNP-1; 312 nm, and SNP-2; 310 nm, Table 1) are attributed to b2 → e3 electronic transitions [17]. Both primary and secondary peaks of SNP-2 are up to 2-3 nm blue-shifted compared to SNP-1, confirming that bare sulfur nanoparticles is slightly smaller in size compared to Ocimum gratissimum stabilized analogue, due to quantum confinements of the later. (Table 1), (Figure 1).
The stabilization of sulfur nanoparticles in the absence (oxalic acid only; SNP-2) and presence (SNP-1) of bioactive molecules of Ocimum gratissimum were confirmed by the FTIR spectra shown in Figure 2A & B. The broad absorption band of SNP-2 centered at 3363 is associated with the –OH stretching of oxalic acid; while the broad and split bands in SNP-1 around 3201-3514 , are associated with the combined –OH and --NH groups of fatty and amino acids in the plant extract [18-20]. This is an indication that the bioactive molecules of Ocimum gratissimum extract act as capping agents on synthesized sulfur nanoparticles. In SNP-1 spectrum, characteristic absorption bands at 2926, 2851 , 1656 , and 1616 , corresponding to , , CONH (amide), and C=C in SNP-1, respectively, which are not present in SNP-2, confirmed the interaction between the protein in the plant extract and the biosynthesized nanoparticles. Thus, FTIR spectrum of SNP-1 differs from that of SNP-2. In SNP-2, intense absorption peaks at 1650 was assigned to carbonyl stretching (C=O) of oxalic acid. The multiple strong bands in the range of 1315–1385 are due to the overtones of C-O stretch (alcohol), C=S stretch (sulfide), and S-O stretch (sulfoxide) [11,21]. Successful formation of sulfur nanoparticles was confirmed by the presence of absorption bands ranging from 550-817 in SNP-1; this peak shifted to 553-840 in SNP-2, due to the interaction of sulfur nanoparticles with the biomolecule contents of Ocimum gratissimum leaf extract [17,22,23]. (Figure 2 A & B)
Morphology of as-prepared sulfur nanoparticles are verified using Scanning Electron Microscope (SEM), Figures 3A & B. The SEM images show that oxalic acid capped sulfur nanoparticles are clusters of small-sized and spherically shaped with definite morphologies. Biosynthesized nanocrystals on the other hand are conspicuously aggregated with irregular particle sizes and shapes, Figure 3B. The EDS spectrum of bare nanoparticles shows the presence of C, O, and Na alongside the S atom peak, Figure 4A. Aside the S peak which confirms the formation of sulfur nanoparticles, the presence of Na, O and C peaks are from and oxalic acid used as precursors for the preparation of sulfur nanoparticles. EDX analysis of biosynthesized sulfur nanoparticles shows the presence of intense S peak, confirming successful formation of sulfur nanoparticles. Other elements observed in the EDX image alongside S peak are Ca, Na, C, O, and Mg, figure 4B. Mineral content analysis of the leaves of Ocimum gratissimum in previous literature reports have shown that they are very rich in Ca, Mg, Na and K [24], hence the presence of Ca and Mg peaks in the EDX image of Ocimum gratissimum stabilized sulfur nanoparticles.
(Figure 3 A & B). (Figure 4 A & B).
Elemental distributions as obtained from the EDX spectra (Figure 4A & B) showed the weight and atomic % of S atoms obtained from SNP-1and SNP-2. Interestingly, at the same approximate concentrations of and oxalic acid, weight and atomic % of S atom in sulfur nanoparticles of SNP-2 is more than two times larger than S atom in SNP-1 prepared from oxalic acid alone. Also, the total weight or atomic % of other mineral ions presence in SNP-2 is far less than that of S atom, suggesting that the prepared nanoparticles composed solely of sulfur. (Figure 5 A & B). Transmission Electron Microscope (TEM) micrograph confirm the synthesized sulfur nanoparticles with oxalic acid (SNP-1) are monodispersed, and spherically shaped particles with average particle size of 18 ± 4 nm. The TEM image of SNP-2 on the other hand showed that the spherically shaped sulphur nanoparticles are aggregated with undefined sizes. The observed dark grey areas in the TEM micrograph of SNP-2are due to the presence of bioorganic molecules from the Ocimum gratissimum leaf extract (Figure 6).
The crystalline nature of the as-synthesized sulfur nanoparticles, in the presence and absence of Ocimum gratissimum leaf extract was analyzed using X ray diffraction patterns in Figure 7. Both sulfur nanoparticles showed similar XRD patterns with ten distinct diffraction peaks located at 0 values of ca. 15.4°, 22.12°, 23.19°, 25.37°, 26.10°, 28.11°, 29.18°, 31.79°, 37.04°, and 43.11°, corresponding to crystal facets of 111, 220, 222, 040, 422, 206, 313, 044, 422, and 319, respectively. The positions, intensities and crystalline phases of as-synthesized sulfur nanoparticles were similar and consistent with the reported standard diffraction patterns of Joint Committee on Powder Diffraction, Standard (JCPDS No. 08247). Average particle sizes of synthesized sulfur nanoparticles are determined by Debye–Scherrer formula, Eqn 2.

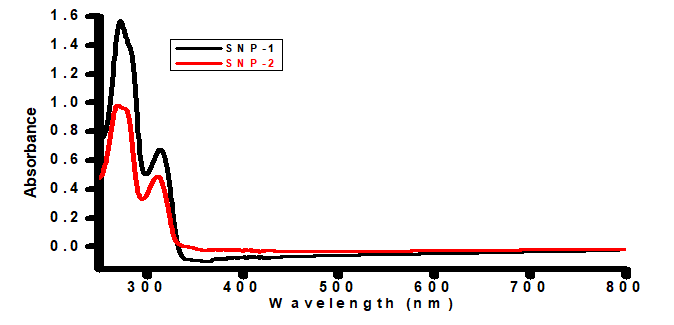
Figure 1: UV-Vis absorption spectra of bare and Ocimum gratissimum capped sulfur nanoparticles.
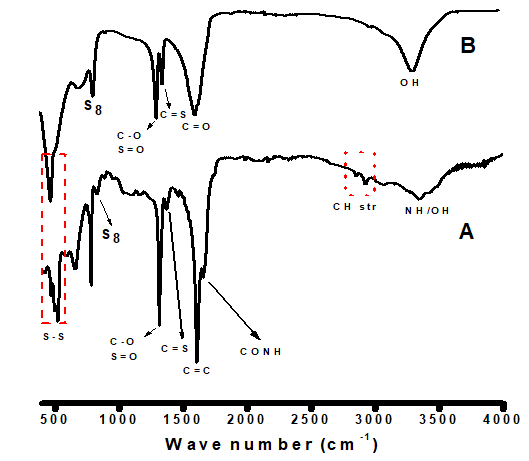
Figure 2 A & B: Fourier Transform Infrared (FTIR) spectroscopy analysis of bare and Ocimum gratissimum capped sulfur nanoparticles.
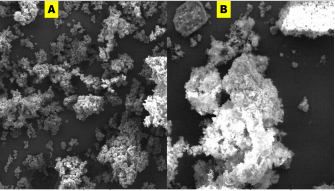
Figure 3 A & B: Scanning electron microscope images of (a) bare, and (b) biosynthesized sulfur nanoparticles.
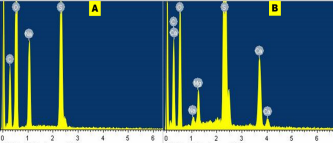
Figure 4 A & B: Energy dispersive spectra of the synthesized (A) bare sulfur nanoparticles, and (B) biosynthesized sulfur nanoparticles.
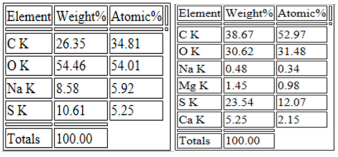
Figure 5: Elemental distribution of of sulfur nanoparticles synthesized in the absence (SNP-1; left) and presence (SNP-2; right) of aqueous leaf extract of Ocimum gratissimum.
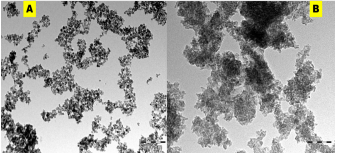
Figure 6: Transmission electron microscope of (A) SNP-1 and (B) SNP-2.
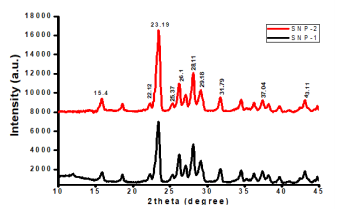
Figure 7: X-RD analysis of sulfur nanoparticles synthesized in the absence (SNP-1) and presence (SNP-2) of aqueous leaf extract of Ocimum gratissimum.
Where D is mean particle size, λ is X-ray wavelength, β is the full width of half maximum of diffraction lines, and θ is the diffraction angle. Average particle size of SNP-1 and SNP-220 and 34 nm, respectively. (Figure 7).
The diameter (in mm) of the zone of inhibition of the sulfur nanoparticles tested against selected clinical isolates. Staphylococcus aureus was observed to have the highest susceptibility to Sulfur Nanoparticles (SNP1) mediated with Ocimum gratissimum plant extract with diameter of 20.00 mm; Escherichia coli and Salmonella pullorum showed the susceptibility diameter zone of 18.00 mm; Klebsiella pneumoniae and Pseudomonas aeruginosa showed resistance. All tested clinical isolates were resistant to the other Sulfur Nanoparticles (SNP2) synthesized in the absence of Ocimum gratissimum plant extract.
Antibacterial efficacy of Ocimum gratissimum capped sulfur nanoparticles against multidrug-resistant microbes (mm)
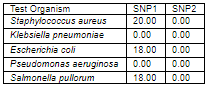
Key: SNP 1 - Sulfur Nanoparticle 1 SNP 2 - Sulfur Nanoparticle 2 0.00 - No inhibition.
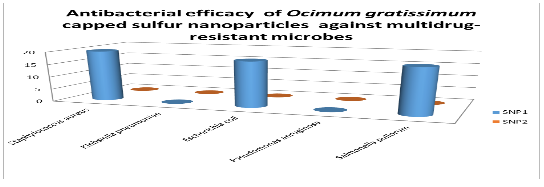
for the clinical isolates; Staphylococcus aureus was observed to have the highest susceptibility to Sulfur Nanoparticles (SNP1) mediated with Ocimum gratissimum plant extract with diameter of 20.00 mm; Escherichia coli and Salmonella pullorum showed the susceptibility diameter zone of 18.00 mm; Klebsiella pneumoniae and Pseudomonas aeruginosa showed resistance. All tested clinical isolates were resistant to the other Sulfur Nanoparticles (SNP2) synthesized in the absence of Ocimum gratissimum plant extract.
The result obtained also showed that Ocimum gratissimum extract increased the efficacy of sulfur nanoparticle as antimicrobial agent. The antimicrobial properties of elemental sulfur have long been recognized, and the use of Sulfur Nanoparticles (SNPs) as antimicrobial agents was first proposed by Lawson (1934). Since then, Schneider et al. (2011) demonstrated that SNPs of about 150 nm in size exhibited antimicrobial activity against various microorganisms. Two sulfur nanoparticles were used in this study (Table 13); SNP1 was mediated with Ocimum gratissimum plant extract while SNP2 was synthesized in the absence of Ocimum gratissimum plant extract. Ocimum gratissimum is commonly called ‘Efinrin nla’ and is widely cultivated in Nigeria for its medicinal use (Aguiyi et al., 2000), Ocimum gratissimum phytochemical screening has indicated the presence of various phytoconstituents which has been shown to be active against several bacteria including Staphylococcus aureus, Listeria monocytogenes, Escherichia coli (Rishikumar et al., 2015; Bankole, 2018). SNP2 showed no antibacterial effect on all the bacteria (endophytic and clinical isolates) used in this study while SNP1 showed potential antibacterial effect on three of the five selected clinical isolates; Staphylococcus aureus were observed to have the highest susceptibility with diameter of 20.00 mm; Escherichia coli and Salmonella pullorum showed the susceptibility diameter zone of 18.00 mm. The endophytic bacterial isolates were also susceptible to the nanoparticles with lower diameter zones ranging from 9mm-14mm. The Sulfur Nanoparticle (SNP1) showed potential antibacterial activity, the amount of nanoparticle used maybe increased to achieve even better outcome. Schneider et al. (2011) demonstrated that SNPs has no toxicity to human cells, thereby making it more considerable as an alternative antimicrobial agent.
References:
- Jeevanandam J, Barhoum A, Chan YS, Dufresne A and Danquah MK. Review on nanoparticles and nanostructured materials: history, sources, toxicity and regulations, Beilstein Nanotechnol J. 2018;9:1050-1074.
- Ogunsona EO, Muthuraj R, Ojogbo E, Valerio O and Mekonnen TH. Engineered nanomaterials for antimicrobial applications: A review, Applied Materials Today, 2020;18:100473-10050.
- Sánchez-Lopez E, Gomes D, Esteruelas G, Bonilla L, Lopez-Machado AL and Galindo R, et al. Metal-Based nanoparticles as nntimicrobial agents: an overview, Nanomaterials. 2020;10:292-331.
- Hoseinzadeh E, Makhdoumi P, Taha P, Hossini H, Stelling J and Kamal MA, et al. Review on nano-antimicrobials: metal nanoparticles, methods and mechanisms, Curr Drug Metab. 2017;18:120-128.
- Yao Y, Zang Y, Qu J, Tang M and Zhang T. The toxicity of metallic nanoparticles on liver: the subcellular damages, mechanisms, and outcomes, International Journal of Nano medicine. 2019;14:8787-8804.
- Salem NM, Albanna LS, Awwad AM, Ibrahim QM and Abdeen AO. Green synthesis of nano-sized sulfur and its effect on plant growth, Journal Agricultural Science, 2016;8:188-194.
- Rai M, Ingle AP and Paralikar P. Sulfur and sulfur nanoparticles as potential antimicrobials: from traditional medicine to nanomedicine, Expert Rev Anti-infect Ther. 2016;14:969-978.
- Paralikar P, Ingle AP, Tiwari V, Golinska P, Dahm H and Rai M. Evaluation of antibacterial efficacy of sulfur nanoparticles alone and in combination with antibiotics against multidrug-resistant uropathogenic bacteria, J. Environ Scientific Health. 2019;54:381-390.
- Shankar S, Pangeni R, Park JW and Rhim JW. Preparation of sulfur nanoparticles and their antibacterial activity and cytotoxic effect, Mater Scientific Engineering. 2018;92:508-517.
- Paralikar P and Rai M. Bio-inspired synthesis of Sulfur nanoparticles using leaf extract of four medicinal plants with special reference to their antibacterial activity, IET Nanobiotechnol. 2017;12:25-31.
- Khairan K, Zahraturriaz and Jalil Z. Green synthesis of sulphur nanoparticles using aqueous garlic extract (Allium sativum), Rasayan Journal of Chemistry. 2019;12:50-57.
- Nweze EI and Eze EE. Justification for the use of Ocimum gratissimum L in herbal medicine and its interaction with disc antibiotics, BMC Complement Altern Med. 2009;9:37-43.
- Kin A, Yaki LM, Abubakar1 I, Olusola LF and Zubairu R. Antibacterial activity of Ocimum gratissimum (scent leaf) on some pathogenic gastrointestinal bacteria, African Journal of Microbiology Research 2018;12:923-929.
- Thakur S, Das G, Raul PK and Karak N. Green one-step approach to prepare sulfur/reduced graphene oxide nanohybrid for effective mercury ions removal, Journal of Physics Chemistry. 2013;117:7636-7642.
- Tripathi RM, Pragadeeshwara Rao R and Tsuzuki T. Green synthesis of sulfur nanoparticles and evaluation of their catalytic detoxification of hexavalent chromium in water, RSC Adv. 2018;8:36345-36352.
- Suryavanshi P, Pandit R, Gade A, Derita M, Zachino S and Rai M, Colletotrichum sp.- mediated synthesis of sulphur and aluminium oxide nanoparticles and it’s in vitroactivity against selected food-borne pathogens, LWT-Food Sci. Technol. 2017;81:188-194.
- Richardson N and Weinberger PJ. Electron Spectrosc. Relat. Phenom. 1975;6:109-116.
- Pratheeba T, Ragavendran C and NatarajanLarvicidal D. pupicidal and adulticidal potential of Ocimumgratissimum plant leaf extracts against filariasis inducing vector, International Journal of Mosquito Research. 2015;2:01-08
- Macdonald IDG and Smith WE. Orientation of cytochrome c adsorbed on a citrate-reduced silver colloid surface, Langmuir. 1996;12:706-713.
- Das B, Dash SK, Mandal D, Ghosh T, Chattopadhyay S and Tripathy S, et al. Green synthesized silver nanoparticles destroy multi drug resistant bacteria via reactive oxygen species mediated membrane damage, Arabian Journal of Chemistry. 2017;10:862-876.
- Vinayan BP, Zhao-Karger Z, Diemant T, Chakravadhanula VSK, Schwarzburger NI and Cambaz MA, et al. Performance study of magnesium-sulfur battery using a graphene based sulfur composite cathode electrode and a non-nucleophilic Mg electrolyte, Nanoscale. 2016;8:3296-3306.
- Davis RE and Nakshbendi HF, Journal of the American Chemistry Society. 1962;84: 2085-2090.
- Prathna T, Chandrasekaran N, Raichur AM and Mukherjee A, Colloids Surf., B, 2011;82:152-159.
- Adebola A. Mineral contents of O. gratissimum leaves (African Basil), International Journal of Science and Research. 2017;6:1572-1573.
- CLSI. Clinical and Laboratory Standards Institute; Wayne, PA: (2009). Methods for dilution antimicrobial susceptibility testing for bacteria that grew aerobically. M7-A10.
- Oludare Temitope Osuntokun, Thonda Oluwakemi A and Adeleye Bukola M. Exploring the Efficacy of Medicinal Plants (Moringa Oleifera and Tamarindus Indica Seeds) in the Treatment of Well Water in Two Major Cities in Southwestern Part of Nigeria, West Africa. Drug Designing & Intellectual Properties International Journal. 2019;3:339-350.
- Lawson GB. The inhibitory action of sulfur on the growth of tubercle bacilli. American Review of Tuberculosis. 1934;29:650-651.
- Aguiyi JC, Obi CI, Gyang SS and Igweh AC. Hypoglycemic activity of Ocimum gratissimum in rats. Fitoterapi. 2000;71:444-446.
- Rajeshkumar S, Malarkodi C, Gnanajobitha G, Paulkumar K, Vanaja M and Kannan C, et al. Seaweed-mediated synthesis of gold nanoparticles using Turbinariaconoides and its characterization. J. Nanostruct. Chem. 2013;3:1-7.
- Schneider T, Baldauf A, Ba LA, Jamier V, Khairan K and Sarakbi MB et al. Selective antimicrobial activity associated with sulfur nanoparticles. Journal of Biomedical Nanotechnology. 2011;7:395-405.
- Bankole OM. Phytochemical and Antimicrobial Studies of Colloidal Silver Nanoparticles Mediated by LaporteaAestuansExtract. International Journal of Biomedical Engineering and Clinical Science. 2018;4:58-65.

