Review of Research Progress on Detection and Identification Methods of Mycobacterium
Guan-Ying Ma, Zhi-Gang Zhang,
Department of Clinical Pathology, Chengde Medical University, China.
Department of Pathology, Cangzhou People’s Hospital, China.
Received Date: 04/12/2021; Published Date: 29/12/2021
*Corresponding author: Zhi-gang Zhang, Department of Pathology, Cangzhou People’s Hospital, 7 Qingchi Avenue, Cangzhou City, Cangzhou 061000, China.
Abstract
Nontuberculosis-Nlycobacteria (NTM), also known as environmental mycobacteria or mycobacteria other than tuberculosis, it grows in both natural and man-made environments and plays an important role in mycobacteria,and some of its strains can cause disease. With the increasing incidence of lung diseases caused by NTM worldwide, NTM has become an important human pathogen, especially in recent years, a number of nosocomial NTM infections have occurred in China, which has gradually aroused concern at home and abroad. The premise of clinically targeted treatment is accurate identification of strains; however, the imaging manifestations of mycobacterium subspecies are not easy to distinguish and the pathomorphologic manifestations are very similar, all of which are granulomatous inflammatory manifestations. At present, the reagent that can detect the most species of bacteria is the gene detection kit of Aneugen Biotechnology (Shenzhen) Co., Ltd., so how to detect more than 22 species of mycobacteria is of great significance. With the increasing incidence of mycobacterial disease, it is urgent and necessary to find a simple and efficient detection method. This paper describes the experimental principles, advantages and disadvantages as well as accuracy of several generations of mycobacterium identification methods, in order to provide support for clinical and laboratory personnel. For example, traditional bacterial identification methods are simple but time-consuming; the method of molecular identification is rapid and specific, but not economical and simple; first-generation sequencing can accurately detect more than 22 strains of mycobacterium. If tuberculosis is not confirmed, metagenomic second-generation sequencing can be used to detect mycobacterium, but the second-generation sequencing is complicated, time-consuming, multi-loci and expensive, so the first-generation sequencing has its limitations but is convenient and quick. With the outbreak of the epidemic, the disease has become the focus of clinical research. Despite the continuous discovery and application of different treatment methods, the diagnosis of mycobacterium still faces great challenges, and there is an urgent need to develop new and efficient identification methods to improve the accuracy. Mycobacteriosis caused by non-tuberculous mycobacteria is an increasingly concerned patient with immune deficiency and immune capacity in human medicine, but there are still many deficiencies in the identification methods of strains. In order to search for an efficient, convenient and economical identification method, this paper describes the new progress in the identification technology of non-tuberculous mycobacteria.
Keywords:Mycobacteria;Traditional bacteriological identification;Metagenomic sequencing;Generation sequencing; Research progress
Introduction
Nontuberculosis Nlycobacteria (NTM), also known as mycobacterium environmental or Mycobacterium extrinsic tuberculosis (MOTT), refers to mycobacteria other than mycobacterium tuberculosis complex and Mycobacterium leprosy [1], which is an important opportunistic pathogen that usually lives freely. Widespread in the environment. So far, about 200 species of NTM have been identified as human pathogens, and they can cause a wide range of infections, with lung infections accounting for 65%-90% of them [2]. Infection of pulmonary NTM is usually caused by primary pulmonary disease, whereas disseminated infections and lymphadenitis caused by NTM are common in immunodeficient hosts [3]. NTM is not a single disease, but a general term for bronchopulmonary diseases caused by any mycobacterium except mycobacterium tuberculosis. It is mainly harmful to people with chronic pulmonary diseases, nosocomial infections and reduced immunity. For example, non-tuberculous mycobacteriosis without clinical symptoms is reported to account for 25% of cases [4]. Although mycobacterium tuberculosis infection accounts for the main part of granulomatous inflammation, there are many patients with clinical symptoms of non-tuberculous mycobacterium, and hospital-acquired infection, such as aubaniere infection of non-tuberculous mycobacterium found in our hospital, neither patient has obvious clinical manifestations. Line two strains of mycobacteria classified spot hybridization experiments show that all the following mycobacteria (shame after mycobacteria, mycobacterium tuberculosis complex group, secondary mycobacteria, intracellular mycobacteria, bird mycobacteria, stomach mycobacteria, Kansas mycobacteria, scrofula mycobacteria, cow mycobacteria, turtle mycobacteria, turtle mycobacterium abscess bacteria, sur plus mycobacteria, Sea/ulcer mycobacteria, toad mycobacteria, di's mycobacteria, accidental/pig mycobacteria, yellow mycobacterium, ape mycobacteria, soil mycobacteria, grass mycobacteria, do not produce color mycobacteria, Gordon mycobacteria), and then to identify the bacteria strains of metagenomic detection are tips for Obama's mycobacteria, This finding also plays a preventive and protective role for medical workers. Most of the clinical manifestations of Mycobacterium are not characteristic, including cough, sputum, fever, shortness of breath, hemoptysis, chest tightness and chest pain. Imaging performance is also very similar, more performance and lobular core nodes, millet tree-in-bud nodules, interlobular septal thickening, large consolidation, bronchiectasis, ground glass density, pleural thickening, lymph node enlargement, such as performance of (Figure 1-4) is difficult to identify [5], but different subspecies of bacterial species treatment difference is very big, Therefore, accurate and rapid identification of bacterial species is the key to the diagnosis and treatment of various mycobacterial infectious diseases. Caused by rare mycobacteria NTM is rarely, if ever, found that the existing clinical experience and knowledge is not enough, with the progress of science and technology, methods for the identification of strains of constant progress, now roughly divided into traditional bacteriological methods such as survey method, molecular biology technology, the development of rapid molecular diagnostic test to identify mycobacterium tuberculosis has become the focus of the research and implementation [6]. Among them, first-generation sequencing (Sanger sequencing) is found to be a convenient, economical and fast identification method after many reviews and summaries of literatures.
Microscopic and CT images of mycobacterium diagnosis:
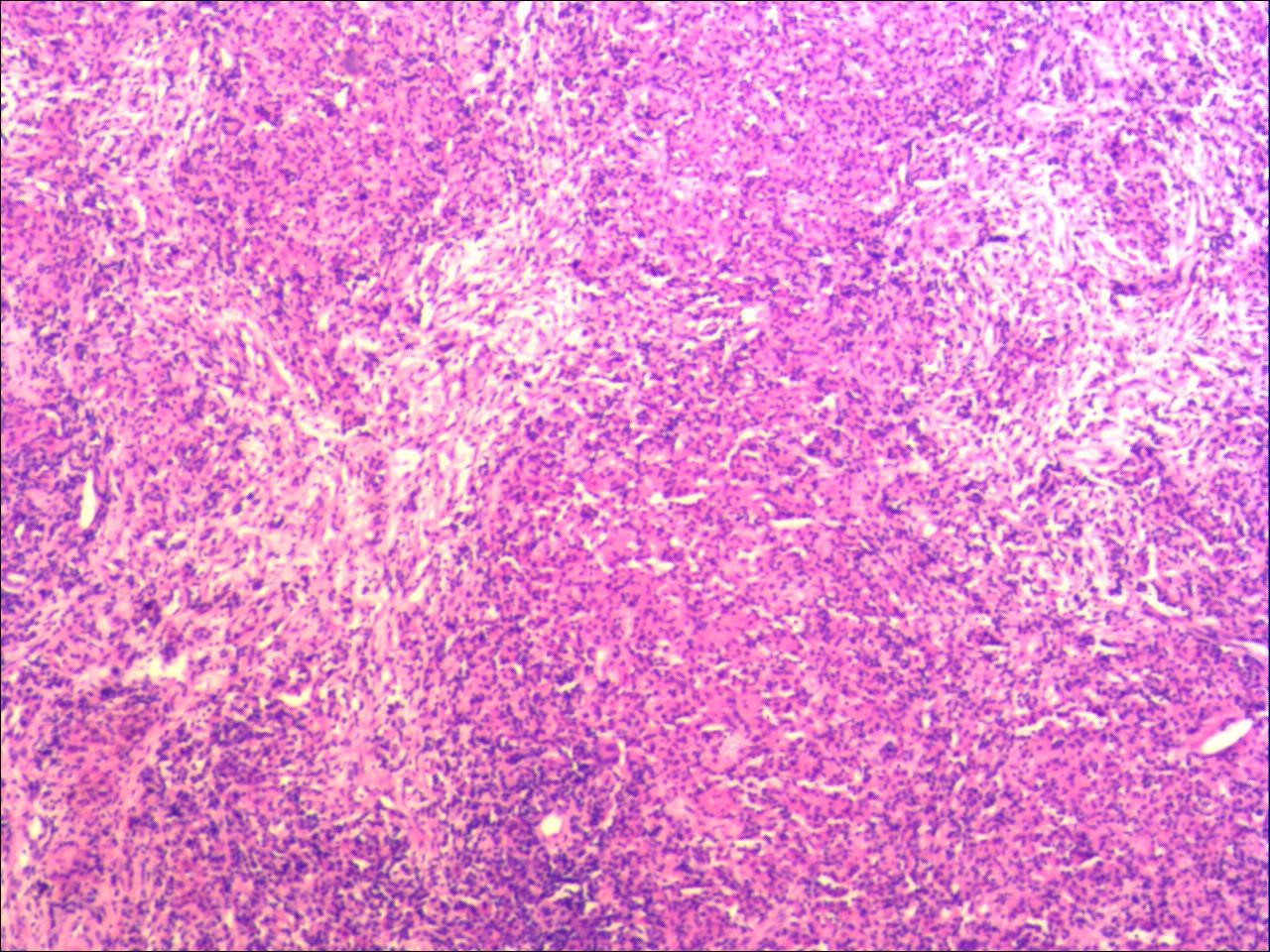
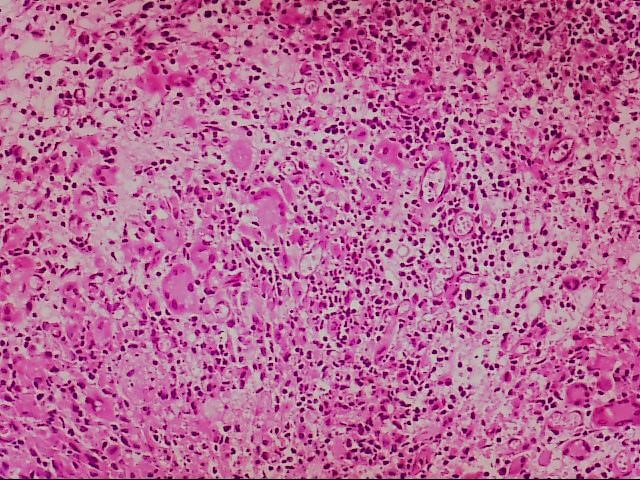
Figure 2:
Microscopically, caseous necrosis, peripheral fibrous connective tissue, and chronic inflammatory cell infiltration are seen

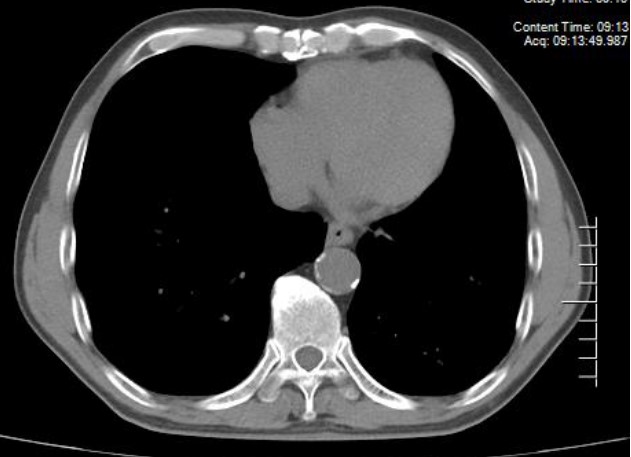
Figure 4
CT mediastinal window showed micronodules, calcification and pleural thickening, ground glass shadow and tree bud sign in lung window.
Traditional Detection Methods for Mycobacteria
Traditional bacteriological identification
Traditional bacteriological identification is mainly based on the morphological, physiological and biochemical characteristics of mycobacteria, including acid-fast staining, fluorescence microscopy, traditional solid culture method, and rapid culture method using semi-automatic culture system, tube culture system, and automatic culture system.
Acid-fast staining and fluorescence microscopy are common staining methods for the detection of mycobacteria. The most commonly used method in the laboratory is the direct smear method of sputum specimens. This method is simple and cheap, but the sensitivity is poor, and the detection rate is not high. The results cannot be obtained within 1 day in the case of a large amount of sputum smear, which is far from meeting the needs of tuberculosis prevention and control. The traditional solid cultivation method of separation takes longer, generally need eight to 12 weeks to get, and the cultivation of the faster technology still need 4-6 weeks 9 [7], and use modified Roche solid culture medium (L - J) culture also needs 20 days to 2 months, even with more advanced automatic mycobacteria/drug susceptibility testing system for liquid culture also needs 7 to 20 days or so. The Capital Bio Real-time Polymerase chain reaction test showed good diagnostic accuracy for both mycobacterium tuberculosis and non-mycobacterium tuberculosis infection. This is of great significance for the differential diagnosis of early pulmonary mycobacterium infection [8].
In recent years, the rapid development of fluorescent staining technology, as a research method or experimental means, has been widely used in the field of medical science in a variety of basic theoretical research and clinical diagnosis. The sensitivity, specificity and accuracy of bacterial assay are much higher than that of bacterial assay. However, isolation culture is the "gold standard" for tuberculosis diagnosis in terms of tuberculosis control, epidemiological investigation and research.
Timed fluorescence quantitative PCR
Timed fluorescent quantitative PCR is a method to measure the total amount of products formed after each cycle of Polymerase chain reaction in DNA amplification reaction using fluorescent chemicals. Because the use of fluorescent chemicals can enable the detection personnel to conduct real-time detection of the Polymerase chain reaction process, PCR sequencing of 16S-23S rDNA [9] is more sensitive, and this gene fragment is generally used to detect cases with low content of acid-fast bacilli and atypical changes in tissue morphology. It can avoid the false negative caused by the limitation of acid-fast staining to a certain extent and improve the detection rate of bacteria. PCR has double specificity including probes and primers. Rapid diagnostic methods based on real-time PCR, such as GeneXpert MTB/RIF method, can detect rifampin (RIF) resistance at the same time. Moreover, it is easy to operate in a closed tube and causes less samples and environmental pollution. However, it also has the disadvantages of easy interference of primers, limited detection types and low amplification efficiency. The establishment of THE LNA-OSN-Q-PCR method can redesign the probe based on the existing mature N-PCR, or redesign the primers based on the mature Q-PCR. Lna-osn-q-pcr does not need to open the lid in the whole process, and has almost no contamination risk. With its high accuracy, good repeatability, quantitative analysis and short time consuming, it can be used as a routine molecular diagnostic method in microbiology laboratories on a large scale, and has the potential to become a commercial kit.
Pcr-Reverse Dot Hybridization (RDBH)
Dot hybridization is to add denatured DNA to nitrocellulose membrane, hybridize with labeled probe, autoradiography, judge whether there is hybridization and hybridization intensity, used for gene deletion or copy number change detection. A new Reverse Dot Hybridization (RDBH) method was developed for rapid identification of clinical mycobacterium isolates. Using 16S rRNA as the target, this method was identified by DNA sequencing and conventional methods including growth characteristics, pigment production, colony morphology and biochemical tests. Data showed that the sensitivity of mycobacterium probe hybridized to the membrane of clinical isolates was 100% [10], which greatly improved the detection efficiency of mycobacterium. But its detection coverage is limited.
Molecular Detection of Mycobacterium
Matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS)
MALDI—TOF-MS mainly produces different track spectra under the action of magnetic field according to the bacterial protein, and compares them with the spectrum in the database to obtain the result of bacterial species identification. The combination of QPCR and MALDI-TOF remains a suitable method for timely diagnosis of mycobacterium species. This may ultimately help detect cases missed by phenotypic testing and enhance NTM diagnostic capabilities to effectively improve patient management [11]. The 16SrRNA gene fragment in molecular detection technology is sensitive, and its main defect lies in that some non-tuberculate mycobacterium species cannot be detected. However, as its relevant database is the most complete at present, it is recommended for routine use, and the data obtained by this method for NTM strain identification are of clinical value and epidemiological significance. Therefore, rapid identification of Mycobacterium tuberculosis complex and NTM is the key to appropriate treatment [12].
Gene chip technology
The common chip technologies are DNA microarray method and Liquid chip (Suspension array). The known sequence amplified by FLUORESCENCE labeling PCR is used for qualitative and quantitative analysis by hybridizing signal with oligonucleotide probe on the chip. The fluxes of the two microarray methods were higher in the detection of drug resistance genes and identification of bacterial strains. Although the identification ability was weaker than that of direct gene sequence analysis, the synthesis of nucleic acid probes was prone to errors or impurities, which reduced the specificity of hybridization. However, this technology is simple to operate, capable of identifying common clinical NTM, and can solve most clinical problems [13]. The whole process of the chip method used in this study only takes 4 hours [14]. After initial screening by chip hybridization, 16S rRNA gene sequencing was selected for confirmation. Although the selected 16S rRNA gene identification ability is relatively low, due to its most complete relevant database at present, it is recommended for routine use [15]. Currently, there are few detection items available for solid-phase chip, so it is difficult to be widely used.
Sanger Sequencing
The clinical radiological characteristics of this disease and tuberculosis (TB) were confirmed and identified by intra transcriptional spacer gene sequencing in 16S-23S rRNA. Patients with active disease were treated by NTM species and followed up to determine clinical, radiological, and microbiological outcomes [16]. With the development of molecular biology technology, DNA sequencing identification method has become the gold standard for bacterial species identification. DNA sequencing identification method for mycobacterium species identification is based on certain specificity of certain homologous DNA fragments of different mycobacteria. This method has high resolution and short detection time. DNA sequence number of clustering analysis is the first to use DNA sequence method for the identification of mycobacteria strains, the appearance of DNA sequence database makes DNA sequence contrast identification of mycobacterium species no longer need to professional and technical personnel can also be measured, some open public information database platform to enrich the database itself, such as America's national centre for biological information (NCBI), However, some scholars found that NCBI has some identification error sequences, incomplete sequences and unnamed sequences, which can affect the accuracy of identification results [17]. Due to the shortcomings of traditional detection methods such as low sensitivity and long time, misdiagnosis or missed diagnosis is easy to be caused to some patients. Therefore, whether to provide rapid, sensitive, specific, reliable, simple and suitable for clinical application of laboratory examination technology has become the focus of research at home and abroad. First-generation sequencing has the advantages of sensitivity, specificity and rapidity, which can improve the detection rate of TUBERCULOSIS bacilli and play an important role in promoting the prevention and treatment of tuberculosis in China and the world.
Metagenomics Next Generation Sequencing (NGS)
NGS can be analyzed by genomics method to extract DNA from the environment, which can cover the entire genome instantly without prior knowledge of specific pathogens. It is suitable for mycobacterium species that are not easy to find, but its accuracy depends on accurate extraction of high concentration OF DNA or RNA and efficient removal of host genes. Although second-generation sequencing can make up for the limitations of traditional identification technology, it is difficult to popularize widely due to its high price, high requirements for identification operation and limited research environment. A rapid, simple, specific and sensitive tuberculosis diagnostic test can accurately identify the existing technology can't detection of mycobacterium tuberculosis strains, the itself patients with lung basic diseases such as tuberculosis, the defects in advance by means of screening for genetic genes or gene screening to regular follow-up, prevent prompt action, it is of great help and significance to clinical diagnosis and treatment [18].
Prospect
China is a country with a high burden of TB, and the burden of non-tuberculous mycobacterium (NTM) disease is also increasing worldwide, but its diagnosis is still delayed and mistaken for multi-drug resistant tuberculosis (MDR-TB) [19] The proportion of diseases caused by non-tuberculous mycobacterium is increasing, long-term ineffective treatment and continuous TB transmission. It seriously affects the prevention, control and management of TUBERCULOSIS and poses a major threat to human health. However, the lack of an organized monitoring system for NTM, such as that for TUBERCULOSIS, still delays its diagnosis and is easily misdiagnosed as multidrug-resistant tuberculosis (MDR-TB) [20].
Existing application of species identification technology, the traditional test method of overall existence one-sided sex, although the sensitivity is high, the operation is simple and the price is low, but time-consuming, specificity and reliability is low, can't close relatives of accurate ruled out identification of strains of mycobacterium tuberculosis, however, because of its low cost, and still is the core of all diagnostic methods, it is widely used in clinical practice. With increasing attention paid to Non-Tuberculous Mycobacterium (NTM) disease, especially in health infection outbreaks, researchers are increasingly developing molecular biotechnology and gene bank testing systems to detect and control NTM at low cost. Maldi-tof-ms [21], PCR amplification and dot hybridization are relatively efficient and reliable identification methods with high resolution, good repeatability and short detection time. These new technologies greatly improve the accuracy of NTM detection. In addition, it has been applied in some large hospitals and laboratories [22]. However, comprehensive data on patient susceptibility, dominant species and drug resistance profile are also needed in order to improve treatment plan and management of NTM. Although only a limited number of antibiotic NTM show sensitivity, the resistance of isolates is highly variable, so more caution should be taken in the empirical treatment of NTM infection [23]. At present, there are still many non-tuberculous mycobacterium disease-causing strains that cannot be identified with model specificity. Therefore, rapid identification of NTM from MTBC and specific identification of NTM are essential for correct treatment and correct patient management.
Therefore, the Sanger sequencing mentioned in this paper can solve the problems of time-consuming, complex operation and low cost. In the future, a simple, rapid, convenient and reliable routine laboratory NTM species identification technology will be established. This method can be used to identify the most common NTM from lung samples in developing countries [24]. Due to the complex identification mechanism of non-tuberculous mycobacteria, which is not completely clear and difficult to determine, there is still a lack of very effective and accurate methods. The above methods have shown the advantages of detection in experimental studies, but more studies are needed to explore the clinical application and specific implementation methods. There is still a long way to go in the research and selection of methods to identify non-tuberculous mycobacteria. It is believed that in the near future, people can conquer the disease caused by non-tuberculous mycobacteria, which seriously affects human health.
References
- Singh Kamal, Kumari Richa, Tripathi Rajneesh, et al. Detection of clinically important non tuberculous mycobacteria (NTM) from pulmonary samples through one-step multiplex PCR assay. BMC Microbiol, 2020, 20
- Nithichanon Arnone, Chetchotisakd Ploenchan, Matsumura Takayuki, et al. Diagnosis of NTM active infection in lymphadenopathy patients with anti-interferon-gamma auto-antibody using inhibitory ELISA vs. indirect ELISA. Sci Rep, 2020, 10: 8968.
- Small PM, Pai M. Tuberculosis diagnosis: time for a game change. N Engl Med, 2010; 363: 1070–1071.
- Ruvandhi R, Nathavitharana, Patrick GT Cudahy, Samuel G. Schumacher, et al. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis [European Respiratory Journal], 2017; 49(1).
- Ko JM, Park HJ, Kim CH, et al. The relation between CT findings and sputum microbiology studies in active pulmonary tuberculosis. Eur J Radiol, 2015; 84(11): 2339-2344.
- Heifets LB, Cangelosi GA. Drug susceptibility testing of Mycobacterium tuberculosis: a neglected problem at the turn of the century. Int J Tuberc Lung Dis, 1999; 3: 564-581.
- Dinnes J, Deeks J, Kunst H, et al. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess, 2007; 11: 1-196.
- Agizew T, Basotli J, Alexander H. Higher-than-expected prevalence of non-tuberculous mycobacteria in HIV setting in Botswana: Implications for diagnostic algorithms using Xpert MTB/RIF assay. PLoS One, 2017;12(12). doi: 10.1371/journal.pone.0189981.
- Ahmad S, Mokaddas E. Diversity of Nontuberculous Mycobacteria in Kuwait: Rapid Identification and Differentiation of Mycobacterium Species by Multiplex PCR, INNO-LiPA Mycobacteria v2 Assay and PCR Sequencing of rDNA. Med Princ Pract, 2019; 28(3): 208-215.
- Kalaiarasan, Ellappan, Thangavelu, Kalpana, Krishnapriya, Krishnakumariamma, et al. Diagnostic performance of real time PCR and MALDI-TOF in the detection of nontuberculous mycobacteria from clinical isolates. Tuberculosis (Edinb), 2020; 125: 101988.
- Arora Disha, Dhanashree Biranthabail, Mycobacterium tuberculosis Utility of smear microscopy and GeneXpert for the detection of in clinical samples. Germs, 2020; 10: 81-87.
- Shen Yanqin, Fang Likui, Xu Xudong, et al. Capital Bio Mycobacterium real-time polymerase chain reaction detection test: Rapid diagnosis of Mycobacterium tuberculosis and nontuberculous mycobacterial infection. Int J Infect Dis, 2020; 98: 1-5.
- Keerthirathne TP, Magana-Arachchi DN, Madegedara D, et a1. BMC Infect Dis. 2016; 16(1): 607.
- Wu Xueqiong, Zhang Junxian, Liang Jianqin, et a1. Comparison of three methods for rapid identification of mycobacterial clinical isolates to the species level. Journal of clinical microbiology, 2007; 45(6).
- Chinese Medical Association Tuberculosis Branch, non-tuberculous mycobacteriosis laboratory diagnosis expert consensus writing group. Expert consensus on laboratory diagnosis of non-tuberculous mycobacteriosis. Chinese journal of tuberculosis and respiration, 2016; 39 (6): 438-443.
- Matsumoto Y,Kinjo T,Motooka D,et a1.Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg Microbes Infect, 2019; 8(1): 1043 1053.
- Sharma SK, Sharma R, Singh BK, et a1. A prospective study of non-tuberculous mycobacterial disease among tuberculosis suspects at a tertiary care centre in north India. Indian J Med Res. 2019; 150(5): 458-467. doi: 10.4103/ijmr.
- Fang H, Shangguan Y, Wang H, et a1. Multicenter evaluation of the biochip assay for rapid detection of mycobacterial isolates in smear-positive specimens. Int J Infect Dis, 2019; 81: 46-51.
- Liu Guan, LI Qi, Huang Hairong. Identification of mycobacterium species by DNA sequencing. Chin J tuberculosis respiratory, 2015; 38(10): 765-767.
- Singh Kamal, Kumari Richa, Tripathi Rajneesh et al. Detection of clinically important non tuberculous mycobacteria (NTM) from pulmonary samples through one-step multiplex PCR assay. BMC Microbiol, 2020; 20: 267.
- Lei Z, Da X, HanCan L, et al. Trends in the Prevalence and Antibiotic Resistance of Non-tuberculous Mycobacteria in Mainland China, 2000-2019: Systematic Review and Meta-Analysis. Front Public Health, 2020; 8: 295.
- kyar I, Cavu§o§lu C, Aya§M, et a1. Evaluation of the per-fofinance of MALDI-TOF MS and DNA sequence analysis in the identification of mycobacteria species. Turk J Med Sci, 2018; 48(6): 1351-1357.
- Xuan Z, Jian L, YuXiang X, TaoSheng Y. Research progress in identification of non-tuberculous mycobacterium species. Chin J tuberculosis lung health, 2019; 8(02): 146-148.
- Singh Kamal, Kumari Richa, Tripathi Rajneesh et al. Detection of clinically important Non-Tuberculous Mycobacteria (NTM) from pulmonary samples through one-step multiplex PCR assay. BMC Microbiol, 2020; 20: 267.

