p66ShcA, A Context Dependent Player in Promoting Cellular Plasticity, Lung Metastasis and an Epigenomic Basis for Breast Cancer Metastasis
Hudson J*
Department of experimental medicine, McGill University, Montreal, Quebec
Received Date: 16/06/2020; Published Date: 20/07/2020
*Corresponding author: Jesse Hudson, Doctor in experimental medicine, McGill University, Montreal, Quebec. E-mail: jesse.hudson@mail.mcgill.ca
Editorial
Metastasis is responsible for 90% of cancer-related deaths and includes efficient and inefficient steps, with the later stages being rate-limiting to successfully establishing metastases. Src homology and collagen A (ShcA) is an adaptor protein that relays extracellular signals by coupling to receptor and cytoplasmic tyrosine kinases to control cell proliferation, survival, invasion, and angiogenesis [1]. The ShcA allele encodes three proteins that originate through alternative promoter usage (p66) or alternate translation initiation (p46, p52) [2]. However, the role of p66ShcA in recurrence is conflicting and poorly understood in breast cancer [3], and even across cancer types, as p66ShcA has been reported to have pro or anti-metastatic features depending on the context [4]. Reactive oxygen species (ROS) are important in the regulation of cell proliferation, apoptosis and cell survival. The major source of ROS in the cell is from the mitochondria and the formation of ROS can also enhance the metastatic potential of breast cancers by perturbing mitochondrial respiration and enhancing migration and/or invasion [5]. Indeed, there had not been any in vivo studies performed in breast cancer elucidating the role of p66ShcA in this setting and we recently set out to answer these questions [6].
Previously, we provided the first in vivo evidence of the role of p66ShcA in ErbB2+ breast cancer in inducing an EMT to promote cellular plasticity and tumor heterogeneity (Figure 1), left panel [6]. Reversible conversion between epithelial and mesenchymal states has been associated with metastasis, increased tumor heterogeneity and resistance to therapy. In TNBC, we recently demonstrated that distinct pools of p66ShcA are necessary for lung metastasis [6]. Both cytoplasmic-p66ShcA and mitochondrial-p66ShcA are necessary at different stages (Figure 1, middle panel). At the earliest stages, cytoplasmic-p66ShcA increases the migratory capacity of TNBC cells, potentially to increase access to the vasculature. During extravasation, cytoplasmic pool aids in the turnover of focal adhesions, increasing their assembly/disassembly, reducing their size and surface area to potentially aid in lung colonization and reactivation of proliferation pathways. These data raise the intriguing possibility that integrins and/or additional cytoskeletal proteins (such as FAK/RhoA/Rac1 which have previously been shown to interact with p66ShcA/ShcA proteins) are involved in p66ShcA-dependent colonization to aid in adhesion/tension-induced mechanotransduction. Mitochondrial-p66ShcA, however, is necessary for increasing CTCs in the bloodstream from the primary site and this correlates with lung metastasis from the primary site. Notably, cytoplasmic-p66ShcA was unable to rescue lung metastatic burden from the primary site compared to breast tumors lacking p66ShcA, indicating mitochondrial p66ShcA is necessary for intravasation or increased survival once in the bloodstream.
Intriguingly, a foundational role for the epigenomic contribution to metastasis has been established, whereby a lack of CpG island methylator phenotype (B-CIMP) correlates with recurrence and reduced survival in breast cancer [7]. Of note, p66ShcA expression is up regulated in both TNBC and “claudin-low” metastatic variants to the lung and liver and basal breast tumors compared to the luminal subtype [6]. In addition, p66ShcA is regulated transcriptionally in TNBC, raising the possibility that p66ShcA may be regulated epigenetically during breast cancer metastasis (Hudson et al. Under Review) indeed, an increase in active chromatin was seen at the p66ShcA promoter in basal tumors and in aggressive metastatic variants. Furthermore, CTCF a known regulator of chromatin was enriched at the p66ShcA promoter and PARP1 regulates p66ShcA expression in these cells. These data support the potential role for an epigenomic program in breast cancer metastasis that may include p66ShcA, a novel promoter of breast cancer metastasis. Hence, p66ShcA may serve as a biomarker of aggressive breast cancers at increased likelihood of metastasis to the lung and liver.
Notably, methylation of CpGs within the CTCF-binding site prevents binding of CTCF, integrating DNA methylation and insulator function in gene regulation [8]. Recently, CTCF was shown to regulate the forehead transcription factor (FOXM1, a key regulator of the cell cycle, DNA damage response and EMT), and a CTCF-FOXM1 axis regulates tumour growth and metastasis in ovarian cancer [9,10]. FOXM1 is frequently overexpressed in cancer and a gene expression signature of FOXM1 predicts the prognosis of luminal breast cancer patients [11]. FOXM1 was also found to co-bind with estrogen receptor alpha to regulate luminal breast cancer. Finally, FOXM1 was shown to promote stemness and radio resistance in glioblastoma and increased mitochondrial function and tumorigenesis through expression of PRX3 in colon CSCs [12]. Hence, these findings integrate various processes including EMT, mitochondrial function, stemness and epigenetic regulation by CTCF to coordinate an epigenetic program that supports metastatic progression. It will be interesting to determine whether a CTCF/p66ShcA-driven axis exists and contributes to metastasis. Furthermore, future studies should delineate the importance of p66-dependent regulation on metastasis, potentially focusing on acetylation and methylation and their associated epigenetic players such as Sirt1 and DMNT 1 and 3b.
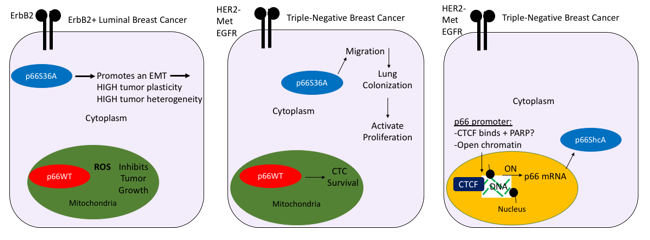
Figure 1: Schematic model of the regulation of and contextual role of p66ShcA in breast cancer across subtypes.
The role of p66ShcA is outlined in an ErbB2+ luminal and basal breast tumor models. Cytoplasmic p66ShcA induces an EMT and promotes cellular plasticity and tumor heterogeneity in a HER+ setting, while cytoplasmic p66ShcA promotes cell migration, colonization of the lung and reactivates cell proliferation pathways at secondary organs (lungs). A schematic of the epigenetic regulation of p66Shca is outlined in TNBC whereby CTCF binds to the p66ShcA promoter at activated chromatin (black marks = active histone marks H3K9Ac and H3K27Ac and beads = Histone 3), potentially through PARP1 or other chromatin modifiers (SIRT1, HATs?) to induce transcriptional expression of p66ShcA in aggressive metastatic breast tumors. Hypomethylated DNA correlates with p66ShcA expression within the p66ShcA promoter and hypomethylated DNA has been proposed as an epigenomic basis of metastasis in breast cancer [7].
References
- Ursini-Siegel J, Hardy WR, Zuo D, Lam SHL, Sanguin-Gendreau V, Cardiff RD, et al. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. The EMBO Journal. 2008;27(6):910-920.
- Ventura A. The p66Shc Longevity Gene Is Silenced through Epigenetic Modifications of an Alternative Promoter. Journal of Biological Chemistry. 2002;277(25):22370-22376. https://doi.org/10.1074/jbc.M200280200.
- Frackelton AR, Lu L, Davol PA, Bagdasaryan R, Hafer LJ, & Sgroi DC. p66 Shc and tyrosine-phosphorylated Shc in primary breast tumors identify patients likely to relapse despite tamoxifen therapy. Breast Cancer Research. 2006;8(6):1-8. https://doi.org/10.1186/bcr1631.
- Li X, Xu Z, Du W, Zhang Z, Wei Y, Wang H, et al. Aiolos promotes anchorage independence by silencing p66Shctranscription in cancer cells. Cancer Cell. 2014;25(5):575-589. https://doi.org/10.1016/j.ccr.2014.03.020.
- Kundu N, Zhang S, & Fulton AM. Sublethal oxidative stress inhibits tumor cell adhesion and enhances experimental metastasis of murine mammary carcinoma. Clinical & Experimental Metastasis. 1995;13(1):16-22. https://doi.org/10.1007/BF00144014.
- Hudson J, Ha JR, Sabourin V, Ahn R, La Selva R, Livingstone J, et al. p66ShcA Promotes Breast Cancer Plasticity by Inducing an Epithelial-to-Mesenchymal Transition. Molecular and Cellular Biology. 2014;34(19):3689-3701. https://doi.org/10.1128/MCB.00341-14.
- Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LGT, et al. Breast Cancer Methylomes Establish an Epigenomic Foundation for Metastasis. Science Translational Medicine Bell and Felsenfeld. 2011.
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482-485. doi:10.1038/35013100.
- Zhao L, Yang Y, Yin S, Yang T, Luo J, Xie R, et al. CTCF promotes epithelial ovarian cancer metastasis by broadly controlling the expression of metastasis-associated genes. Oncotarget. 2017;8(37):62217-62230. https://doi.org/10.18632/oncotarget.19216.
- Song B, Chu I. A gene expression signature of FOXM1 predicts the prognosis of hepatocellular carcinoma. Exp Mol Med. 2018;50:e418. https://doi.org/10.1038/emm.2017.159.
- Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013;14(1):R6. Doi:10.1186/gb-2013-14-1-r6.
- Zhang J, Chen XY, Huang KJ, et al. Expression of FoxM1 and the EMT-associated protein E-cadherin in gastric cancer and its clinical significance. Oncol Lett. 2016;12(4):2445-2450. doi:10.3892/ol.2016.4917.

