The Role of KMT2A Gene Amplification in Acute Myeloid Leukemia, Serie of Cases and Literature Review
López de la Osa María José1,*, Juárez-Salcedo Luis Miguel1, Arranz Eva1, Ortiz Javier1, Benzo Gonzalo1, Cannata Jimena1, Loscertales Javier1, Alegre Adrián1 and Dalia Samir2,*
1Hematology Department, La Princesa University Hospital, Madrid, Spain
2Hematology/Oncology Department, Mercy Clinic Oncology and Hematology, Joplin, Missouri, United States
Received Date: 10/01/2025; Published Date: 12/02/2025
*Corresponding author: López de la Osa María José, Hematology Department, La Princesa University Hospital, Madrid, Spain; Dalia Samir, Hematology/Oncology Department, Mercy Clinic Oncology and Hematology, Joplin, Missouri, United States
Abstract
Despite lysine methyl transferase 2A (KMT2A) gene rearrangement representing a common oncogenic event in acute myeloid leukemia (AML), KMT2A amplification is less frequent and is associated with distinct clinic and genetic features. We present a retrospective analysis of three patients with AML associated with KMT2A gene amplification and we realized a literature review. All cases are men, with a median age of 65 years. Two of them had received previous therapy. The bone marrow aspirate smears revealed a marked dysplasia with cytoplasmic vacuolation in erythroid series. In all cases, the cytogenetic study showed highly complex karyotypes and fluorescence in situ hybridization (FISH) analysis showed del (5q) and deletion of the TP53 gene in all patients and del(7q) in two patients. Next-generation sequencing (NGS) panel was performed in two patients, in these cases, biallelic alteration of TP53 was established. All patients were refractory to therapy and the median survival was 74 days after KMT2A amplification appeared. In summary, our results suggest that our patients with KMT2A amplification share the same clinical and genetic characteristics described in the literature: the presence of the amplification in patients with previous chemotherapy, advanced age, signs of dysplasia with frequent vacuolation, complex karyotype, and TP53 mutation.
Keywords: KMT2A gene; Gene amplification; Acute myeloid leukemia; TP53 mutation
Introduction
The lysine methyl transferase 2A (KMT2A) / Mixed Lineage Leukemia (MLL) gene, located on chromosome 11q23, encodes a histone methyl transferase that is crucial in the epigenetic regulation of transcription. It is necessary for the normal development of hematopoiesis and its deregulation leads to the development of leukemia [1-3].
Although balanced rearrangements involving the KMT2A gene have been widely described in AML, KMT2A gene amplification (≥ 4 copies) is a rare condition reported in approximately 1% of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Most AML cases with KMT2A amplification have been associated with advanced age, previous therapy, and adverse outcomes. KMT2A amplification was closely related to highly complex karyotype, losses affecting 5 or 5q (-5/5q-) and TP53 loss/mutation. Moreover, published data suggest that amplification of KMT2A has an important role in leukemogenesis and probably its pathogenesis is different from KMT2A rearrangement [4,5].
We describe detailed clinical, morphological and genetic features of three cases of AML with KMT2A amplification and a literature review to deepen the knowledge of the pathogenesis of this rare genetic abnormality.
Case Presentation
Patient 1
A 78-year-old man with a previous history of radiotherapy for prostate cancer five years earlier. He presented asthenia with laboratory findings of hemoglobin 8.1g/dl, leukocytes 6,150 x 109, and platelets 21 x 109/l. The bone marrow (BM) aspirate smears showed erythroid series with dysplastic signs, binucleated cells, and abundant vacuolations; the granulocytic series showed dysplastic signs with abundant immature forms; 38% of blasts with vacuolations were observed (Figure 1A).
The immunophenotype of blast cells presented monocytic differentiation. He had a hypodiploidy highly complex karyotype (Table 1) and fluorescence in situ hybridization (FISH) analysis showed KMT2A gene amplification with 4 to 14 copies (Figure 2), deletion of TP53 gene and deletion of 5q31, 5q33, 7q22 and 7q31 chromosomal regions. A Next-generation sequencing (NGS) panel was not performed.
The patient received azacytidine presenting nephrotoxicity on the fourth day so it was stopped. He finally presented septic shock with poor evolution and finally died 42 days after diagnosis.
Patient 2
A 65-year-old man without a previous history of treatment. He consulted to emergency department for spontaneous bleeding and bruising. The laboratory analysis showed Hb 11.9g/dl, leukocytes 2440 x 109/l, and platelets 18,000 x 109/l. The bone marrow (BM) aspirate showed an erythroid series with marked dysplasia and the presence of vacuolations; the granulocytic series showed signs of dysplasia; 24% of blasts were observed (Figure 1B).
The immunophenotypic analysis showed two populations of blasts with myeloid differentiation (MPO+, CD117+, CD13+, CD33++, CD123+, CD35-, CD64-, CD14-, CD11b, CD38+, CD71+), one of them with lower complexity showed too CD34+, CD117++, DR+, CD15-. He presented hypodiploidy complex karyotype including der(5;17) (p10;q10) translocation (Table 2), and FISH analysis showed KMT2A gene amplification with 1 to 10 copies, deletion p53 gene, and deletion 5q31 chromosomal region. NGS panel showed mutation of TP53 and DNMT3A.
He received treatment with Vyxeos® and was refractory after two cycles, finally died 6 months after diagnosis.
Patient 3
A 60-year-old man with a previous history of multiple myeloma (MM) IgG kappa four years earlier; was treated with Bortezomib-Dexamethasone and an autologous stem cell transplantation (ASTC) for consolidation followed by lenalidomide maintenance. Routine blood tests showed: Hb 11 g/dl, leukocytes 1480 x 109/L, and platelets 28,000 x 109/l. With these findings, a BM aspirate was requested and showed dysplasia of the erythroid and granulocytic lineages with 20% of blasts. Vacuoles were not observed.
The immunophenotypic analysis showed 32% of blast blasts with myeloid differentiation (CD117+, CD33+, CD13+, HLA-DR+, CD45+). He had a complex karyotype including der(5;17) (p10;q10) translocation (table 2) and the FISH study showed deletion of the TP53 gene and deletion of 5q31, 5q33, 7q22, and 7q31 chromosomal region. At that time KMT2A amplification was not observed. NGS panel showed TP53 mutation.
He was treated with two cycles of Vyxeos®, remaining in complete remission and azacitidine maintenance until receiving allogeneic Hematopoietic Stem Cell Transplantation (HSCT).
In the reassessment of BM aspirate before HSCT, he presented dysplasia of erythroid and granulocytic series, and 10% of blasts vacuolated (Figure 1C). He persisted with a complex karyotype and FISH showed the previous alterations and KMT2A gene amplification de novo with 2 to 6 copies. NGS panel at that moment was not performed. Finally, the patient died on day +44 of HSCT due to toxicity, only 74 days after KMT2A gene amplification appeared.
Table 1 shows the main clinical and genetic characteristics of the patients.
Table 1: Characteristics of patients.
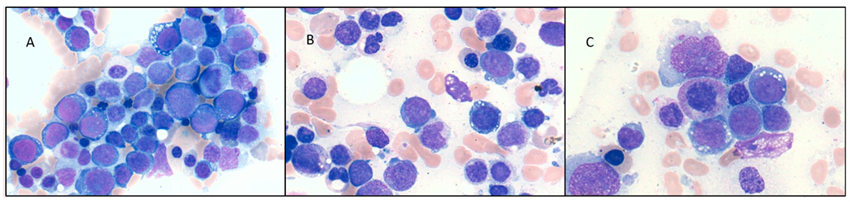
Figure 1: Vacuolated blasts of patients.
Table 2: Karyotypes of patients.


Figure 2: FISH analysis of case 1.
Discussion
The KMT2A gene, located on chromosome 11q23, encodes a large histone methyltransferase that positively regulates gene transcriptions including Homebox (HOX) genes. HOX genes are transcriptional regulators essential during embryogenesis and their regulation is fundamental for tissue development including the hematopoietic system. KMT2A is expressed in most tissues, including myeloid cells and it is important for the maintenance of HOX gene expression and this is necessary for the proliferation and survival of hematopoietic stem cells (HSC). The most common type of KMT2A gene alteration is the reciprocal translocation, with > 130 partner genes forming chimeric fusion protein in acute lymphoid leukemia (ALL), MDS, and AML. [6,7].
Genomic amplification leads to the inappropriate activation of oncogenes expression on one or more oncogenes located in the amplicon. KMT2A gene amplification is an uncommon event reported in approximately 1% of AML and MDS, we can observe different forms of amplification by FISH: intrachromosomal amplification (HSR), extrachromosomal amplification (double minutes chromosomes), and ring chromosomes. [9]. Data published suggest that KMT2A overexpression results in gain on KMT2A function, KMT2A transcript was indirectly demonstrated by up-regulation of HOXA9 gene expression [8]. The main difference between KMT2A reciprocal translocation and KTM2A amplification is that the former results in the production of novel chimeric proteins with partner genes and the latter results in increasing KMT2A copy number or transcriptional activity [6-9].
We show 3 cases with KMT2A gene amplification that shared similar features. The previous data published described this alteration in older and previously treated patients [5,6-13]. In our series, the median age was 65 years (range 60 – 78). Two patients had received previous therapy; patient 1 received only radiotherapy and patient 3 received a proteasome inhibitor (Bortezomib), an alkylating agent (Melfalan) during the ASCT and an immunomodulator (Lenalidomide). The association between KMT2A gene amplification and radiotherapy or alkylating agents has been previously described in other series. [4,10,12].
The BM aspirate smears found marked dysplasia sing mainly in erythroid series with cytoplasmic vacuolation, with a median of vacuolated blast at diagnosis 24% (range 20 – 38%). Cytoplasmic vacuoles on blast had been described in other series and it seemed to be restricted to the cells with KMT2A amplification [10]. Patient 3 presented this vacuolization at the moment that KMT2A gene amplification appeared, this reinforces the close association.
In all patients, a cytogenetic study presented complex karyotypes and other chromosomal abnormalities commonly detected in complex karyotypes (del 5q, del 17p, del 7q) (4, 10). FISH analysis showed KMT2A gene amplification ranging from 4 to 14 copies. NGS panel was performed in two patients, case 2 showed T53 and DNMT3A mutation, and case 3 panel showed TP53 mutation.
Complex karyotype, losses affecting chromosomes 5 or/and 7, and TP53 deletions/mutations have been associated with KMT2A gene amplification in all previous series [4, 8–13]. Interestingly, patient 3 presented TP53 mutation seven months before the appearance of KMT2A amplification, this evolution has been described previously in three patients in another study [10]. This cytogenetic evolution suggests that mainly TP53 alteration may generate an instability genomic that predisposes to KMT2A amplification.
Another fact of interest is the unbalanced translocation der(5;17)(p10;q10) found in two patients (case 2 and case 3) has not been before associated with KMT2A amplification. Published data suggest that this aberration results in deletions of chromosomes 5q and 17p and is postulated that TP53 is deleted as a result of the translocation der(5;17). [14].
TP53 gene has an important role in maintaining genomic stability and integrity, and mutational inactivation of TP53 has been shown to produce gene [10]. As far as we know, there is only 1 reported case in literature in which TP53 was unaltered [12].
In our series, all patients were refractory to treatment and died with a median survival of 74 days (range 42 days 6 months) after KMT2A amplification emerging, similar to that described in the literature with the median of survival around 1 month [4,6-13]. In one of the largest series, higher KMT2A copies were associated with shorter survival, but more studies are need to clarify if the number of copies plays a role in the poor prognosis [10].
Despite the common features previously described in all patients with KMT2A amplification, the poor prognosis in the majority of these patients is entirely consistent with cytogenetic characteristics, so it is difficult to establish whether KMT2A amplification is only an epiphenomenon of genetic instability or plays a role in leukemia progression.
Conclusion
In summary, KMT2A gene amplification in AML is associated with previous therapy, older age, and aggressive clinical course. KMT2A amplification is typically related to complex karyotype and deletion/mutation of TP53. Our results are consistent with those reported in other series.
Conflicts of interest: The authors declare no conflicts of interest.
References
- Yip BH, Tsai CT, Rane JK, Vetharoy W, Anguita E, Dong S, et al. Amplification of mixed lineage leukemia gene perturbs hematopoiesis and cooperates with partial tandem duplication to induce acute myeloid leukemia. Haematologica, 2017; 102(8): e300-e304.
- Yip BH, So CW. Mixed lineage leukemia protein in normal and leukemic stem cells. Exp Biol Med (Maywood), 2013; 238(3): 315-323.
- Koka R, Mainor CB, Banerjee A, Baer MR, Zou YS. Concomitant amplification of the MLL gene on a ring chromosome and a homogeneously staining region (hsr) in acute myeloid leukemia: mechanistic implications. Leuk Lymphoma, 2017; 58(5): 1250-1253.
- Andersen MK, Christiansen DH, Kirchhoff M, Pedersen-Bjergaard J. Duplication or amplification of chromosome band 11q23, including the unrearranged MLL gene, is a recurrent abnormality in therapy-related MDS and AML, and is closely related to mutation of the TP53 gene and to previous therapy with alkylating agents. Genes Chromosomes Cancer, 2001; 31(1): 33-41.
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet, 2002; 30(1): 41-47.
- Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol, 2012; 7: 283-301.
- Marschalek R. The reciprocal world of MLL fusions: A personal view. Biochim Biophys Acta Gene Regul Mech, 2020; 1863(7): 194547.
- Poppe B, Vandesompele J, Schoch C, Lindvall C, Mrozek K, Bloomfield CD, et al. Expression analyses identify MLL as a prominent target of 11q23 amplification and support an etiologic role for MLL gain of function in myeloid malignancies. Blood, 2004; 103(1): 229-235.
- Cuthbert G, Thompson K, McCullough S, Watmore A, Dickinson H, Telford N, et al. MLL amplification in acute leukaemia: a United Kingdom Cancer Cytogenetics Group (UKCCG) study. Leukemia, 2000; 14(11): 1885-1891.
- Tang G, DiNardo C, Zhang L, Ravandi F, Khoury JD, Huh YO, et al. MLL gene amplification in acute myeloid leukemia and myelodysplastic syndromes is associated with characteristic clinicopathological findings and TP53 gene mutation. Hum Pathol, 2015; 46(1): 65-73.
- Pajuelo-Gámez JC, Cervera J, García-Casado Z, Mena-Durán AV, Valencia A, Barragán E, et al. MLL amplification in acute myeloid leukemia. Cancer Genet Cytogenet, 2007; 174(2): 127-131.
- Zatkova A, Merk S, Wendehack M, Bilban M, Muzik EM, Muradyan A, et al. AML/MDS with 11q/MLL amplification show characteristic gene expression signature and interplay of DNA copy number changes. Genes Chromosomes Cancer, 2009; 48(6): 510-520.
- Maitta RW, Cannizzaro LA, Ramesh KH. Association of MLL amplification with poor outcome in acute myeloid leukemia. Cancer Genet Cytogenet, 2009; 192(1): 40-43.
- Warnstorf D, Bawadi R, Schienke A, Strasser R, Schmidt G, Illig T, et al. Unbalanced translocation der(5;17) resulting in a TP53 loss as recurrent aberration in myelodysplastic syndrome and acute myeloid leukemia with complex karyotype. Genes Chromosomes Cancer, 2021; 60(6): 452-445.

