First Case of the D3a Sub-Genotype Coxsackievirus A6-Associated Hand, Foot and Mouth Disease in Senegal
Khadim Diop1,#, Ndack Ndiaye2,#, Al Ousseynou Seye2,#, Abdou Diop3, Babacar Ndiaye3, Mame Téné Ndiaye1, Emilienne Perronne Paglan1, Ndeye Marieme Ndiaye1, Ousmane Faye4, Moussa Diallo5, NDongo Dia2, Amadou Alpha Sall2 and Martin Faye2,*,#
1Dermatology Pediatric Department, Albert Royer Hospital, Dakar, Senegal
2Virology Department, Institut Pasteur de Dakar, 36 Avenue Pasteur, 220 Dakar, Senegal
3Biomedical Laboratory, Institut Pasteur de Dakar, 36 Avenue Pasteur, 220 Dakar, Senegal
4Department of Public Health, Institute Pasteur de Dakar, 36, Avenue Pasteur, 220 Dakar, Senegal
5Dermatopathology Department, University Cheikh Anta Diop, Dakar, Senegal
#These authors participated equally to this publication
Received Date: 01/10/2024; Published Date: 15/11/2024
*Corresponding author: Martin Faye, Virology department, Institut Pasteur de Dakar, 36, Avenue Pasteur, 220 Dakar, Senegal
Abstract
Hand, Fot and Mouth Disease (HFMD), has been emerging as a common human disease since 2008 in children and adults worldwide. However, the disease seems to be neglected in Africa, where specific surveillance and studies are warranted. This report describes a case of hand, foot and mouth disease in a child, associated with Coxsackievirus A6 in Senegal.
Keywords: Rash; Enterovirus; Coxsackievirus A6; Children; Senegal; Africa; 2024
Introduction
Hand, Foot and Mouth Disease (HFMD) is a viral infection, affecting mostly children under 10 years [1]. Symptoms of HFMD include fever, mouth sores and a skin rash on the hands and feet. Outbreaks of HFMD have been previously reported in many parts of the world and most common causative agents are Coxsackievirus A16 (CVA16), Coxsackievirus A6 (CVA6) and the enterovirus A71 (EV-A71), belonging to the Enterovirus genus (EV) of the Picornaviridae family [2]. However, the clinical features of CVA6-associated HFMD are slightly different from those caused by other enteroviruses. In fact, infections with CVA6, often presents with atypical clinical symptoms including more severe and extensive dermatologic manifestations on the trunk, neck, legs, and perioral area, and onychomadesis which causes nail loss [3]. The CVA6 presents a large diversity with four genotypes designated as A, B, C, and D according to previous data and further subdivided into B1–B2, C1–C2, and D1–D3 sub-genotypes [4]. The D3 sub-genotype was further subdivided into two evolutionary branches including D3a and D3b [4]. In Senegal, HFMD is not a notifiable disease and there is no active surveillance implemented. Herein, we report the first case of CVA6’s sub-genotype D3a-associated HFMD in an eight-month girl admitted at the dermatology department of the Albert Royer pediatric hospital in Dakar, Senegal.
Materials and Methods
Ethical Consideration
The Senegalese National Ethical Committee of the MoHSA approved the surveillance protocol as a piece of research with less than minimal risk, and written consent forms were not required. Oral consent was obtained from the patient’s parents, as required by the Senegalese National Ethical Committee of the MoHSA. The sample was de-identified before we performed virus characterization and analysis.
Case Presentation
On 1 June 2024, an eight-months girl with vesiculobullous rash was admitted at the dermatology department of the Albert Royer pediatric hospital in Dakar, Senegal. She was presented without any pathological findings, with a good psychomotor development. However, on examination, the patient was febrile, had fever with 38°C and a vesiculobullous rash sudden onset, evolving over 3 days. The dermatological examination revealed tense vesiculobullous with small erythematous localized on the forearms, palms and dorsum of the hands, legs and feet. In addition, vesicles at the perioral level and desquamative erythema of the seat were also observed (Figure 1A).
The histological aspect of skin biopsy from the bulla showed significant oedema of the dermal papillae raising the epidermis which contained large vesicles. Moreover, it was associated with ballooned and necrotic keratinocytes, with foci of parakeratosis on the surface. The adjacent dermis contained a moderate infiltrate consisting of lymphocytes and neutrophils (Figure 1B).
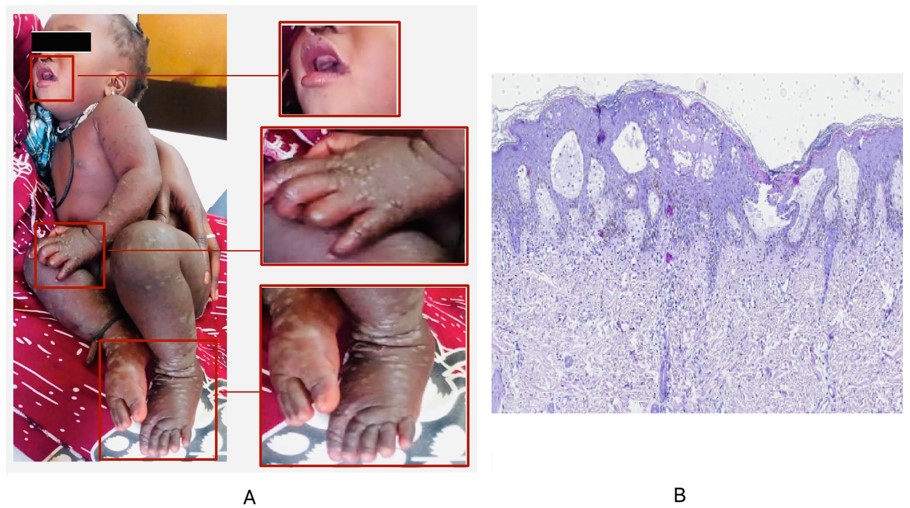
Figure 1: Clinical features of the HFMD case included in our study which shows the appearance of vesicular skin lesions (A) and skin biopsy specimen showing large vesicles with necrotic keratinocytes; œdeme papillae with moderate infiltrate consisting of lymphocytes and neutrophils (H&E, ×100) (B).
Sample collection, RNA extraction and Molecular detection
A swab of the vesicles was collected and shipped to the institute Pasteur de Dakar (IPD) for diagnostic and sequencing, using a virus transportation medium (VTM) (COPAN Diagnostics Inc., Murrieta, CA) at +4°C. Demographic and clinical data were also collected.
Viral RNA was extracted from 200 µL of sample using the QIAmp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The RNA was eluted in 60 μL of nuclease-free water. Then the extracted RNA from the sample was tested by real-time quantitative RT-PCR (qRT-PCR) assay using specific primers and probes for the detection of enteroviruses (EV) [5].
Amplification of the Enterovirus Capsid protein, sequencing and genome analysis
An amplicon of the entire coding-region of the Capsid protein was generated using the OneTaq One-Step RT-PCR kit (New England Biolabs Inc.) with a specific PCR method as previously described [6] and including a negative control containing nuclease-free water. For genotyping, the amplicons were purified using AMPure XP magnetic beads and quantified using the dsDNA High Sensitivity Kit on a Qubit 3.0 fluorometer (Thermo Fisher). Thereafter, the purified DNA were barcoded using the Rapid Barcoding Kit (SQK.110.96) with the MRT001 expansion module (Oxford nanopore technology) and pooled in a single tube. The libraries were then purified and sequenced on a GridIon instrument (Oxford nanopore technology).
Passed reads were analyzed using the metagenomic pipeline implemented on the “Chan Zuckerberg ID - Detect & Track Infectious Diseases'' platform (https://czid.org/) and the obtained sequence was analyzed using the online Basic Local Alignment Search (BLAST) program (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on July 05, 2024) to compare the sequence homology with previously available data. In addition, the genotype was confirmed using the online Enterovirus Genotyping Tool (RIVM) program (enterovirus/typingtool/, accessed on July 05, 2024).
Phylogenetic analysis
Multiple alignments of our dataset including the newly characterized sequence and previously available sequences from Genbank (https://www.ncbi.nlm.nih.gov/), were performed using the BioEdit software (version 7.2.5) [7]. Maximum likelihood (ML) phylogenetic trees based on sequences of the VP1 and the Capsid protein, were inferred using IQ-TREE (version 1.6.12) [8] for 1,000 replications, with the GTR+G find as the best fitted nucleotide substitution model to our dataset. The ML tree’s topology was visualized with Fig Tree (version 1.4.4) [9]. Nodes were supported by Bootstrap values.
Results
Laboratory testing
The swab from the vesicles tested positive for EV with a threshold cycle (Ct) value of 22. A near-complete sequence of the capsid region was obtained with a length of 3,251 nucleotides. After BLAST analysis, the newly characterized complete genome is closely related (96% nt similarity) to a CVA6 strain identified from a child with hand foot and mouth disease from France in 2017 (accession number MT814423.1). The genotyping data were confirmed by the BLAST results.
The ML phylogenetic trees based on sequences of the entire coding-region of the Capsid protein (Figure 2A) and the VP1 protein (Figure 2B), showed that the newly characterized CVA6 sequence clustered with several strains previously identified in the Asia and European countries from 2015 to2018. The new CVA6 sequence from Senegal exhibited a 100% nucleotide homology with an isolate from France in 2017 and belonged to the recently described D3a sub-genotype (Figure 2B).
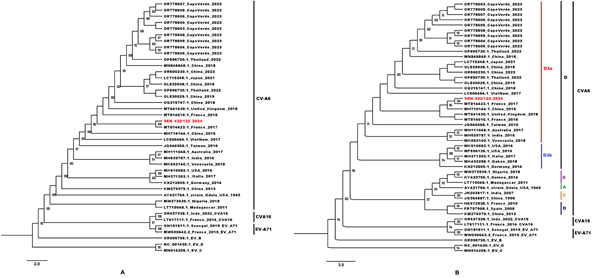
Figure 2: Phylogenetic comparisons trees based on (A) 42 sequences of the entire coding-region of the Capsid (≈3251 pb) and (B) 49 VP1 sequences (≈700 pb) from the newly characterized CVA6 and previously available sequences obtained from GenBank. The newly characterized sequence is highlighted in red. Bootstrap values ≥ 90 are shown on the tree. The scale bar indicates the distances of the branches.
Treatment and evolution
The admitted patient was treated using oral paracetamol, local chlorhexidine-based care and a local healing spray. The evolution was favorable after three and seven days, marked by skin desquamation at day 3 and complete healing of the lesions with residual hypochromia in places at day 7, respectively. There was no recurrence at the two-month visit (Figure 3).
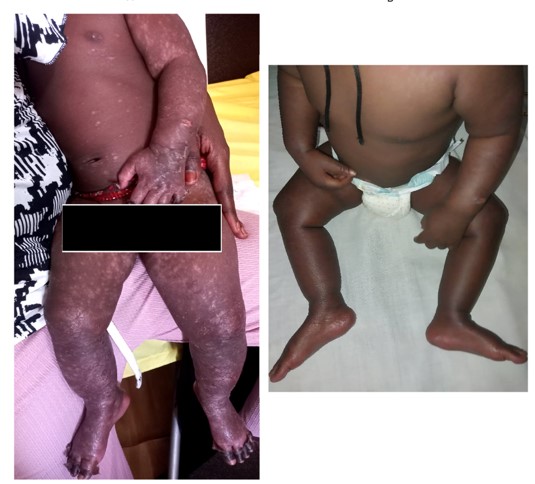
Figure 3: Clinical evolution of the HFMD case after three (A) and seven days (B).
Discussion
HFMD is a common, usually mild childhood illness caused by enteroviruses. Major causes of HFMD include CVA16, CVA6 and EV-A71 [10]. However, EV-A71 is generally responsible for severe cases [10]. In 2018, data monitoring in many Chinese cities has shown that the pathogenic composition of HFMD has changed immensely due to the successful use of the EVA71 vaccine. Thus, CVA6 became the main causative agent of HFMD [11]. Despite the reported circulation of EV-A71, CVA16 and CVA6 in Africa, there is a scarcity of data on the prevalence of HFMD and its causative agents.
In this study, we reported on the clinical presentation and evolution of a CVA6-associated HFMD in an 8-month girl from Senegal.
Children less than five years-old, are the major age group affected by HFMD as previously reported in several studies focusing on HFMD [12].
The case was detected in June which corresponds to the beginning of the summer in Senegal. The period between April and July was previously associated with a high prevalence of HFMD in some parts of China and Japan [13]. In fact, environmental factors such as the average temperature and other prevailing weather conditions could have promoted the highest circulation rates of EV during the summer [13]. However, further longitudinal studies are needed for assessment of the potential seasonal circulation of EV-associated HFMD in Senegal as previously reported in the Shandong region of China [14].
In addition, the role of CVA6 in other diseases could be also investigated as it has been previously reported etiology for meningitis, encephalitis, and acute flaccid paralysis [15].
Interestingly, the new characterized CVA6 sequence exhibited a high nucleotide identity with a CVA6 strain from France in 2017, suggesting a probably importation from Europe. However, due to the lack of sequences from other African countries, the virus circulation dynamic in the region. Thus, regional studies focusing on the genomic surveillance of EV-associated HFMD could be promoted for preparedness.
Our data provide new insights into the molecular epidemiology of CVA6 in Senegal by reporting on the first identification of the D3a sub-genotype which was predominant and co-circulated with other CVA6 genotypes in China from 2017 to 2019 [4]. It was responsible for an Outbreak of HFMD in children from the Cape Verde islands in 2023 (data not shown). Although CVA6-associated HFMD usually heals in seven to 10 days [1], it’s characterized by a high transmissibility [4].
Conclusion
Our study is noteworthy by reporting for the first time on the clinical presentation and evolution of a CVA6-associated HFMD in Senegal. Our data point to the crucial need to estimate prevalence the disease’s prevalence in the country. Therefore, an active hospital-based surveillance of HFMD including genomics could be implemented for rapid identification of emerging EV of public health concern.
References
- Esposito S, Principi N. Hand, foot and mouth disease: current knowledge on clinical manifestations, epidemiology, aetiology and prevention. European Journal of Clinical Microbiology & Infectious Diseases, 2018; 37: 391-398.
- Knowles NJ, Hovi T, Hyypiä T, et al. Family-Picornaviridae, 2012.
- Gaunt E, Harvala H, Österback R, et al. Genetic characterization of human coxsackievirus A6 variants associated with atypical hand, foot and mouth disease: a potential role of recombination in emergence and pathogenicity. The Journal of general virology, 2015; 96(Pt 5): 1067.
- Song Y, Zhang Y, Ji T, et al. Persistent circulation of Coxsackievirus A6 of genotype D3 in mainland of China between 2008 and 2015. Scientific reports, 2017; 7(1): 5491.
- Gunson RN. The Development, Implementation and Evaluation of a Real-Time PCR-Based Diagnostic Service for Viral Causes of Infectious Intestinal Disease. PhD Thesis. University of Glasgow, 2008.
- Majumdar M, Martin J. Detection by direct next generation sequencing analysis of emerging enterovirus D68 and C109 strains in an environmental sample from Scotland. Frontiers in microbiology, 2018; 9: 1956.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium Series. Oxford, 1999; 41: 95-98.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular biology and evolution, 2015; 32(1): 268-274.
- Rambaut A. FigTree, a graphical viewer of phylogenetic trees (tree. bio. ed. ac. 691 uk/software/figtree). Inst Evol Biol Univ Edinburgh, 2014; 1: 2.
- Drago F, Ciccarese G, Gasparini G, et al. Contemporary infectious exanthems: an update. Future Microbiology, 2017; 12(2): 171-193. doi:10.2217/fmb-2016-0147.
- Yu F, Zhu R, Jia L, et al. Sub-genotype change and recombination of coxsackievirus A6s may be the cause of it being the predominant pathogen for HFMD in children in Beijing, as revealed by analysis of complete genome sequences. International Journal of Infectious Diseases, 2020; 99: 156-162.
- Wang Y, Zhao H, Ou R, et al. Epidemiological and clinical characteristics of severe hand-foot-and-mouth disease (HFMD) among children: a 6-year population-based study. BMC Public Health, 2020; 20(1): 801. doi:10.1186/s12889-020-08961-6.
- Yang L, Liu T, Tian D, et al. Non-linear association between daily mean temperature and children’s hand foot and mouth disease in Chongqing, China. Scientific Reports, 2023; 13(1): 20355.
- Zhu P, Ji W, Li D, et al. Current status of hand-foot-and-mouth disease. J Biomed Sci, 2023; 30(1): 15. doi:10.1186/s12929-023-00908-4.
- Machado RS, Tavares FN, Sousa Jr IP. Global landscape of coxsackieviruses in human health. Virus Research, 2024; 344: 199367.

